6UK1
 
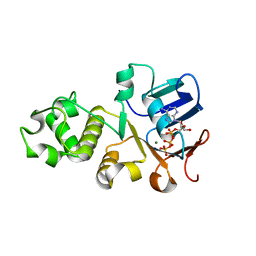 | | Crystal structure of nucleotide-binding domain 2 (NBD2) of the human Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Wang, C, Vorobiev, S.M, Vernon, R.M, Khazanov, N, Senderowitz, H, Forman-Kay, J.D, Hunt, J.F. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | A thermodynamically stabilized form of the second nucleotide binding domain from human CFTR shows a catalytically inactive conformation
To Be Published
|
|
5TF8
 
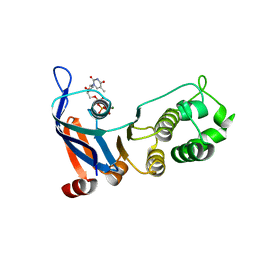 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with dTTP | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-24 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | Thermodynamic correction of F508del-CFTR by ligand binding to a remote site in the mutated domain
To Be Published
|
|
5TFJ
 
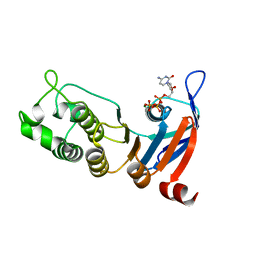 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-25 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Thermodynamic correction of F508del-CFTR by ligand binding to a remote site in the mutated domain
To Be Published
|
|
6KOL
 
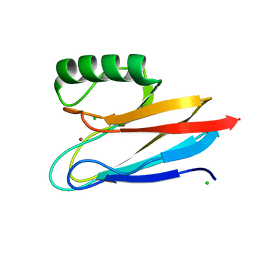 | | Crystal structure of auracyanin from photosynthetic bacterium Roseiflexus castenholzii | | Descriptor: | Blue (Type 1) copper domain protein, CHLORIDE ION, COPPER (II) ION | | Authors: | Wang, C, Zhang, C.Y, Min, Z.Z, Xin, Y.Y, Xu, X.L. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.211 Å) | | Cite: | Structural basis underlying the electron transfer features of a blue copper protein auracyanin from the photosynthetic bacterium Roseiflexus castenholzii.
Photosyn. Res., 143, 2020
|
|
6L9S
 
 | | Crystal structure of Na-dithionite reduced auracyanin from photosynthetic bacterium Roseiflexus castenholzii | | Descriptor: | Blue (Type 1) copper domain protein, COPPER (I) ION | | Authors: | Wang, C, Zhang, C.Y, Min, Z.Z, Xu, X.L. | | Deposit date: | 2019-11-10 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis underlying the electron transfer features of a blue copper protein auracyanin from the photosynthetic bacterium Roseiflexus castenholzii.
Photosyn. Res., 143, 2020
|
|
3PR6
 
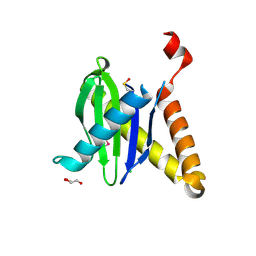 | |
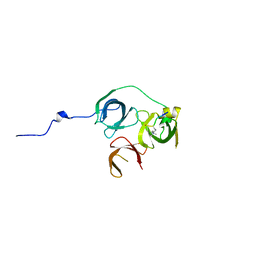
5Y5W
 
 | |
7UVP
 
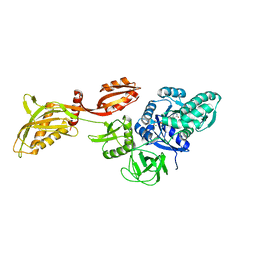 | |
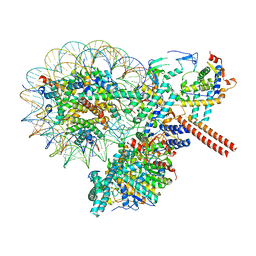
8W9E
 
 | |
8W9F
 
 | |
8W9C
 
 | |
8W9D
 
 | |
4EV8
 
 | |
4EVA
 
 | |
8WX8
 
 | |
4EVT
 
 | |
4EV9
 
 | |
4EVP
 
 | |
8I02
 
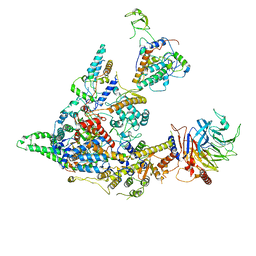 | | Cryo-EM structure of the SIN3S complex from S. pombe | | Descriptor: | Chromatin modification-related protein eaf3, Cph1, Histone deacetylase clr6, ... | | Authors: | Wang, C, Guo, Z, Zhan, X. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Two assembly modes for SIN3 histone deacetylase complexes.
Cell Discov, 9, 2023
|
|
8I03
 
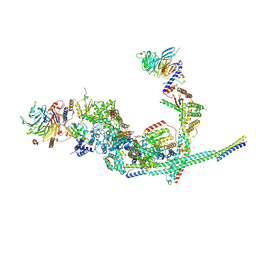 | | Cryo-EM structure of the SIN3L complex from S. pombe | | Descriptor: | Chromatin modification-related protein png2, Histone deacetylase clr6, POTASSIUM ION, ... | | Authors: | Wang, C, Guo, Z, Zhan, X. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Two assembly modes for SIN3 histone deacetylase complexes.
Cell Discov, 9, 2023
|
|
9BRZ
 
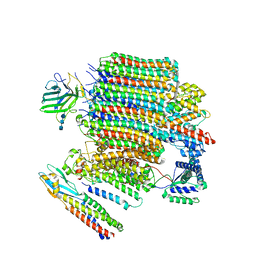 | | V0-only V-ATPase and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
4QIN
 
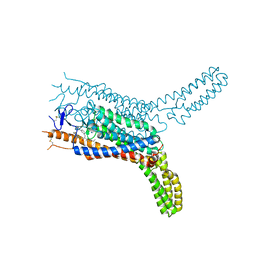 | | Structure of the human smoothened receptor in complex with SAG1.5 | | Descriptor: | 3-chloro-4,7-difluoro-N-[trans-4-(methylamino)cyclohexyl]-N-[3-(pyridin-4-yl)benzyl]-1-benzothiophene-2-carboxamide, Smoothened homolog/Soluble cytochrome b562 chimeric protein | | Authors: | Wang, C, Wu, H, Evron, T, Vardy, E, Han, G.W, Huang, X.-P, Hufeisen, S.J, Mangano, T.J, Urban, D.J, Katritch, V, Cherezov, V, Caron, M.G, Roth, B.L, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2014-05-31 | | Release date: | 2014-07-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs.
Nat Commun, 5, 2014
|
|
9BRU
 
 | | Intact V-ATPase State 1 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRT
 
 | | Intact V-ATPase State 1 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRA
 
 | | Intact V-ATPase State 2 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
