8T1R
 
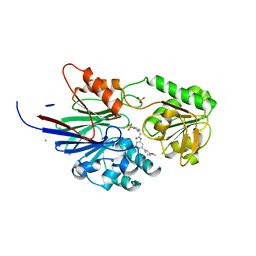 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 2 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(3-methoxyphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
8T1Q
 
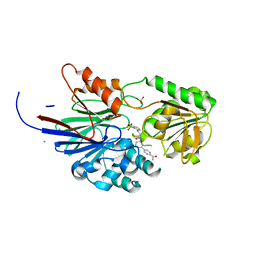 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 1 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(4-ethanoylphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
7EMN
 
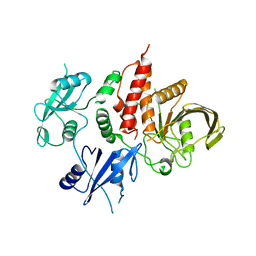 | | The atomic structure of SHP2 E76A mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Luo, F, Xie, J.J, Zhu, J.D, Liu, C. | | Deposit date: | 2021-04-14 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A novel partially open state of SHP2 points to a "multiple gear" regulation mechanism.
J.Biol.Chem., 296, 2021
|
|
4RL3
 
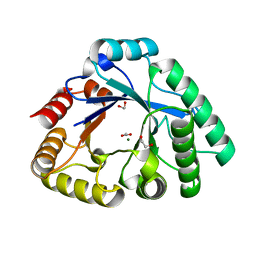 | | Crystal Structure of the Catalytic Domain of a family GH18 Chitinase from fern, Peteris ryukyuensis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Chitinase A, ... | | Authors: | Umemoto, N, Numata, T, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2014-10-15 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A class III chitinase without disulfide bonds from the fern, Pteris ryukyuensis: crystal structure and ligand-binding studies.
Planta, 242, 2015
|
|
8HUB
 
 | | AMP deaminase 2 in complex with an inhibitor | | Descriptor: | 3,3-dimethyl-4-(phenylmethyl)-2~{H}-quinoxaline-1-carboxamide, AMP deaminase 2, ZINC ION | | Authors: | Adachi, T, Doi, S. | | Deposit date: | 2022-12-23 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The discovery of 3,3-dimethyl-1,2,3,4-tetrahydroquinoxaline-1-carboxamides as AMPD2 inhibitors with a novel mechanism of action.
Bioorg.Med.Chem.Lett., 80, 2023
|
|
8HU6
 
 | | AMP deaminase 2 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, AMP deaminase 2, SULFATE ION, ... | | Authors: | Adachi, T, Doi, S. | | Deposit date: | 2022-12-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The discovery of 3,3-dimethyl-1,2,3,4-tetrahydroquinoxaline-1-carboxamides as AMPD2 inhibitors with a novel mechanism of action.
Bioorg.Med.Chem.Lett., 80, 2023
|
|
2EO7
 
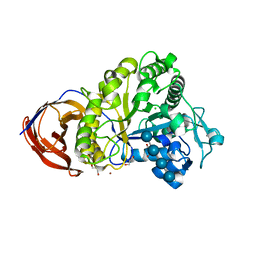 | | Crystal structure of Cel44A, GH family 44 endoglucanase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CHLORIDE ION, Endoglucanase, ... | | Authors: | Kitao, Y, Karita, S, Watanabe, N, Sakka, K, Tanaka, I. | | Deposit date: | 2007-03-29 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum.
J.Biol.Chem., 282, 2007
|
|
7FD6
 
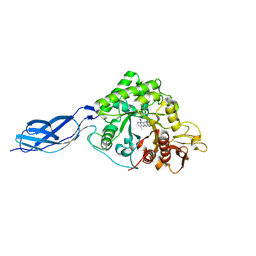 | | Crystal structure of SmChiA in complex with 6a | | Descriptor: | 11-methyl-3-(pyridin-3-ylmethyl)-4H-pyrimido[5'',4'':5',6']pyrido[2',3':4,5]pyrimido[1,2-b]pyridine-4,6(3H)-dione, Chitinase A | | Authors: | Xi, J, Qing, Y. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of Conformation Constrained Tetracyclic Compounds as potent Chitinase OfChi-h inhibitor with novel binding mode
To Be Published
|
|
4EAY
 
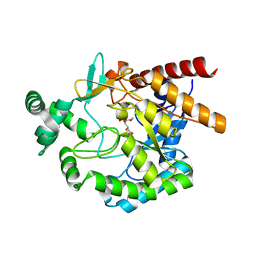 | | Crystal structures of mannonate dehydratase from Escherichia coli strain K12 complexed with D-mannonate | | Descriptor: | CHLORIDE ION, D-MANNONIC ACID, MANGANESE (II) ION, ... | | Authors: | Qiu, X, Zhu, Y, Yuan, Y, Zhang, Y, Liu, H, Gao, Y, Teng, M, Niu, L. | | Deposit date: | 2012-03-23 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into decreased enzymatic activity induced by an insert sequence in mannonate dehydratase from Gram negative bacterium.
J.Struct.Biol., 180, 2012
|
|
4MC5
 
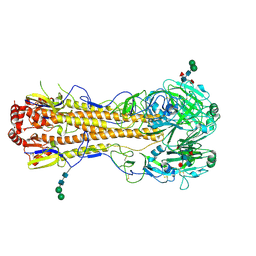 | | Crystal structure of a subtype H18 hemagglutinin homologue from A/flat-faced bat/Peru/033/2010 (H18N11) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Stevens, J. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.238 Å) | | Cite: | New world bats harbor diverse influenza a viruses.
Plos Pathog., 9, 2013
|
|
4MC7
 
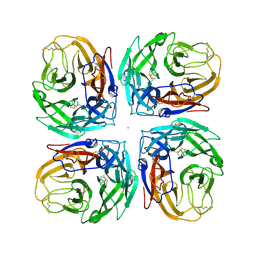 | | Crystal structure of a subtype N11 neuraminidase-like protein of A/flat-faced bat/Peru/033/2010 (H18N11) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Stevens, J. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | New world bats harbor diverse influenza a viruses.
Plos Pathog., 9, 2013
|
|
7XJX
 
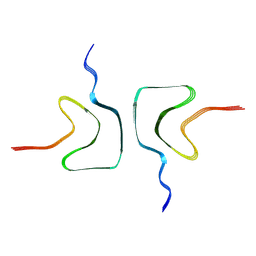 | | The cryo-EM structure of Fe3+ induced alpha-syn fibril. | | Descriptor: | Alpha-synuclein | | Authors: | Zhao, Q.Y, Tao, Y.Q, Zhao, K, Tao, Y.Q, Li, D. | | Deposit date: | 2022-04-18 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural Insights of Fe3+ Induced alpha-synuclein Fibrillation in Parkinson' Disease
J.Mol.Biol., 435, 2023
|
|
8JJ8
 
 | | Cryo-EM structure of the beta2AR-mBRIL/1b3 Fab/Glue complex with a partial agonist | | Descriptor: | Beta-2 adrenergic receptor,Soluble cytochrome b562, ~{N}-[5-[(1~{R})-2-[[(2~{R})-1-(4-methoxyphenyl)propan-2-yl]amino]-1-oxidanyl-ethyl]-2-oxidanyl-phenyl]methanamide | | Authors: | He, B.B, Zhong, Y.X, Guo, Q, Tao, Y.Y. | | Deposit date: | 2023-05-29 | | Release date: | 2023-09-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8JJL
 
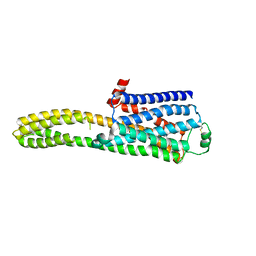 | | cryo-EM structure of the beta2-AR-mBRIL/1b3 Fab/Glue complex with a full agonist | | Descriptor: | Beta-2 adrenergic receptor,Soluble cytochrome b562, Olodaterol | | Authors: | He, B.B, Zhong, Y.X, Guo, Q, Tao, Y.Y. | | Deposit date: | 2023-05-30 | | Release date: | 2023-09-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8J7E
 
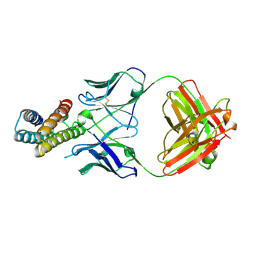 | | Crystal structure of BRIL in complex with 1b3 Fab | | Descriptor: | Antibody 1b3 Fab Heavy chain, Antibody 1b3 Fab Light chain, Soluble cytochrome b562 | | Authors: | Zhong, Y.X, Guo, Q, Tao, Y.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2023-09-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8JJO
 
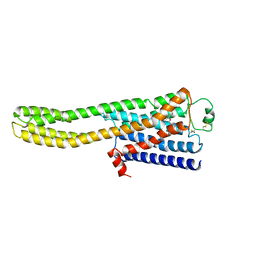 | | Cryo-EM structure of the beta2AR-mBRIL/1b3 Fab/Glue complex with an antagonist | | Descriptor: | (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol, Beta-2 adrenergic receptor,Beta-2 adrenergic receptor,Soluble cytochrome b562 | | Authors: | He, B.B, Zhong, Y.X, Guo, Q, Tao, Y.Y. | | Deposit date: | 2023-05-31 | | Release date: | 2023-09-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
4M6W
 
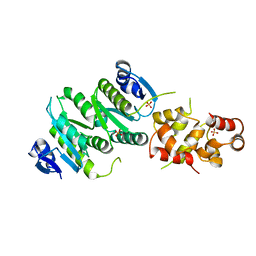 | | Crystal structure of the C-terminal segment of FANCM in complex with FAAP24 | | Descriptor: | Fanconi anemia group M protein, Fanconi anemia-associated protein of 24 kDa, SULFATE ION | | Authors: | Yang, H, Zhang, T, Tong, L, Ding, J. | | Deposit date: | 2013-08-11 | | Release date: | 2013-10-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the functions of the FANCM-FAAP24 complex in DNA repair.
Nucleic Acids Res., 41, 2013
|
|
8ZLI
 
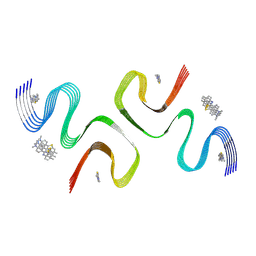 | | BTA-2-bound E46K alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein, ~{N},~{N}-dimethyl-4-(6-methyl-1,3-benzothiazol-2-yl)aniline | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2024-05-20 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8ZLO
 
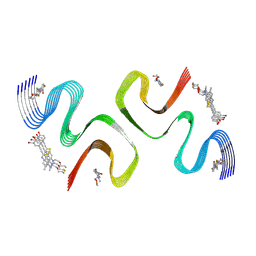 | | F0502B-bound E46K alpha-synuclein fibril | | Descriptor: | 2-bromanyl-4-[(~{E})-2-[6-[2-(2-fluoranylethoxy)ethyl-methyl-amino]-5-methyl-1,3-benzothiazol-2-yl]ethenyl]phenol, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2024-05-20 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8ZMY
 
 | | F0502B-bound WT polymorph 5a alpha-synuclein fibril | | Descriptor: | 2-bromanyl-4-[(~{E})-2-[6-[2-(2-fluoranylethoxy)ethyl-methyl-amino]-5-methyl-1,3-benzothiazol-2-yl]ethenyl]phenol, Alpha-synuclein | | Authors: | Liu, K.E, Tao, Y.Q, Li, D, Liu, C. | | Deposit date: | 2024-05-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Binding adaptability of chemical ligands to polymorphic alpha-synuclein amyloid fibrils.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8ZLP
 
 | |
4QND
 
 | | Crystal structure of a SemiSWEET | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Chemical transport protein, ... | | Authors: | Yan, X, Yuyong, T, Liang, F, Perry, K. | | Deposit date: | 2014-06-17 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Structures of bacterial homologues of SWEET transporters in two distinct conformations.
Nature, 515, 2014
|
|
7XO2
 
 | |
7XO1
 
 | |
7XO0
 
 | |
