2WXH
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with SW14. | | Descriptor: | 2-{[4-amino-3-(3-fluoro-4-hydroxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]methyl}-5-methyl-3-(2-methylphenyl)quinazolin-4(3H)-one, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
2WXK
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with INK666. | | Descriptor: | 3-(2-amino-1,3-benzothiazol-6-yl)-1-{[2-(4-methylpiperazin-1-yl)quinolin-3-yl]methyl}-1H-pyrazolo[3,4-d]pyrimidin-4-amine, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
2WXF
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with PIK-39. | | Descriptor: | 2-((9H-PURIN-6-YLTHIO)METHYL)-5-CHLORO-3-(2-METHOXYPHENYL)QUINAZOLIN-4(3H)-ONE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
2WXO
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with AS5. | | Descriptor: | N-(3-{[(1Z)-3,5-DIMETHOXYCYCLOHEXA-2,4-DIEN-1-YLIDENE]AMINO}QUINOXALIN-2-YL)-4-FLUOROBENZENESULFONAMIDE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
2X6J
 
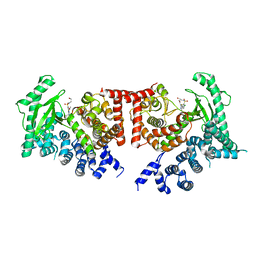 | | THE CRYSTAL STRUCTURE OF THE DROSOPHILA CLASS III PI3-KINASE VPS34 IN COMPLEX WITH PIK-93 | | Descriptor: | N-(5-(4-CHLORO-3-(2-HYDROXY-ETHYLSULFAMOYL)- PHENYLTHIAZOLE-2-YL)-ACETAMIDE, PHOSPHOTIDYLINOSITOL 3 KINASE 59F | | Authors: | Miller, S, Tavshanjian, B, Oleksy, A, Perisic, O, Houseman, B.T, Shokat, K.M, Williams, R.L. | | Deposit date: | 2010-02-17 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Shaping Development of Autophagy Inhibitors with the Structure of the Lipid Kinase Vps34.
Science, 327, 2010
|
|
2WXJ
 
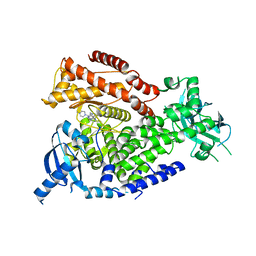 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with INK654. | | Descriptor: | N-[6-(4-amino-1-{[2-(4-methylpiperazin-1-yl)quinolin-3-yl]methyl}-1H-pyrazolo[3,4-d]pyrimidin-3-yl)-1,3-benzothiazol-2-yl]acetamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
2X6K
 
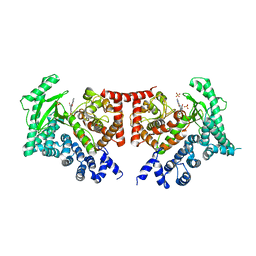 | | THE CRYSTAL STRUCTURE OF THE DROSOPHILA CLASS III PI3-KINASE VPS34 IN COMPLEX WITH PI-103 | | Descriptor: | 3-(4-MORPHOLIN-4-YLPYRIDO[3',2':4,5]FURO[3,2-D]PYRIMIDIN-2-YL)PHENOL, PHOSPHOTIDYLINOSITOL 3 KINASE 59F, SULFATE ION | | Authors: | Miller, S, Tavshanjian, B, Oleksy, A, Perisic, O, Houseman, B.T, Shokat, K.M, Williams, R.L. | | Deposit date: | 2010-02-17 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Shaping Development of Autophagy Inhibitors with the Structure of the Lipid Kinase Vps34.
Science, 327, 2010
|
|
2X6F
 
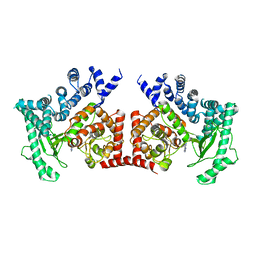 | | THE CRYSTAL STRUCTURE OF THE DROSOPHILA CLASS III PI3-KINASE VPS34 IN COMPLEX WITH 3-METHYLADENINE | | Descriptor: | 6-AMINO-3-METHYLPURINE, PHOSPHOTIDYLINOSITOL 3 KINASE 59F | | Authors: | Miller, S, Tavshanjian, B, Oleksy, A, Perisic, O, Houseman, B.T, Shokat, K.M, Williams, R.L. | | Deposit date: | 2010-02-17 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Shaping Development of Autophagy Inhibitors with the Structure of the Lipid Kinase Vps34.
Science, 327, 2010
|
|
2WXL
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with ZSTK474. | | Descriptor: | 2-(difluoromethyl)-1-(4,6-dimorpholin-4-yl-1,3,5-triazin-2-yl)-1H-benzimidazole, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
3BHI
 
 | |
8T4V
 
 | | Crystal structure of compound 1 bound to K-Ras(G12D) | | Descriptor: | 4-{(1R,5S)-3-[(7P)-7-(8-ethynylnaphthalen-1-yl)-8-fluoro-2-{[(4s,7as)-tetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-4-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl}-4-oxobutanoic acid, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Z, Zheng, Q, Guiley, K.Z, Shokat, K.M. | | Deposit date: | 2023-06-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Strain-release alkylation of Asp12 enables mutant selective targeting of K-Ras-G12D.
Nat.Chem.Biol., 20, 2024
|
|
7E5E
 
 | | Crystal structure of GDP-bound GNAS in complex with the cyclic peptide inhibitor GD20 | | Descriptor: | CHLORIDE ION, GD20, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hu, Q, Dai, S, Shokat, K.M. | | Deposit date: | 2021-02-18 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | State-selective modulation of heterotrimeric G alpha s signaling with macrocyclic peptides.
Cell, 185, 2022
|
|
4IHP
 
 | | Crystal structure of TgCDPK1 with inhibitor bound | | Descriptor: | 1-tert-butyl-3-(3-chlorophenoxy)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1, UNKNOWN ATOM OR ION | | Authors: | El Bakkouri, M, Tempel, W, Crandall, I.E, Massad, T, Loppnau, P, Graslund, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kain, C.K, Shokat, K.M, Sibley, L.D, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-19 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of TgCDPK1 with inhibitor bound
TO BE PUBLISHED
|
|
4J8M
 
 | | Aurora A in complex with CD532 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[[4-[(5-cyclopentyl-1H-pyrazol-3-yl)amino]pyrimidin-2-yl]amino]phenyl]-3-[3-(trifluoromethyl)phenyl]urea, Aurora kinase A, ... | | Authors: | Meyerowitz, J.G, Gustafson, W.C, Shokat, K.M, Weiss, W.A. | | Deposit date: | 2013-02-14 | | Release date: | 2014-09-10 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Drugging MYCN through an Allosteric Transition in Aurora Kinase A.
Cancer Cell, 26, 2014
|
|
4L8G
 
 | | Crystal Structure of K-Ras G12C, GDP-bound | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-06-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
4J8N
 
 | |
4L9W
 
 | | Crystal Structure of H-Ras G12C, GMPPNP-bound | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-06-18 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
4L9S
 
 | | Crystal Structure of H-Ras G12C, GDP-bound | | Descriptor: | CALCIUM ION, GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-06-18 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
4LGH
 
 | |
4LGG
 
 | | Structure of 3MB-PP1 bound to analog-sensitive Src kinase | | Descriptor: | 1-tert-butyl-3-(3-methylbenzyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Lopez, M.S, Dar, A.C, Shokat, K.M. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure-guided inhibitor design expands the scope of analog-sensitive kinase technology.
Acs Chem.Biol., 8, 2013
|
|
4M1Y
 
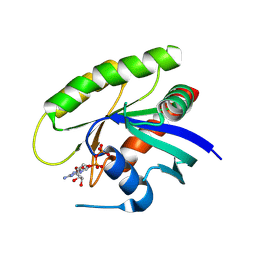 | | Crystal Structure of small molecule vinylsulfonamide 15 covalently bound to K-Ras G12C | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, K-Ras GTPase, N-{1-[N-(5,7-dichloro-2,1,3-benzothiadiazol-4-yl)glycyl]piperidin-4-yl}ethanesulfonamide | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-08-04 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
4LYH
 
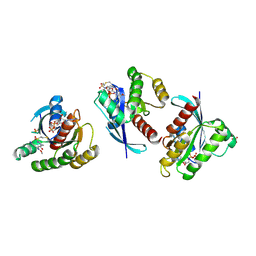 | | Crystal Structure of small molecule vinylsulfonamide 9 covalently bound to K-Ras G12C | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, N-{1-[N-(4-chloro-5-iodo-2-methoxyphenyl)glycyl]piperidin-4-yl}ethanesulfonamide, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-07-31 | | Release date: | 2013-11-27 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.371 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
4M22
 
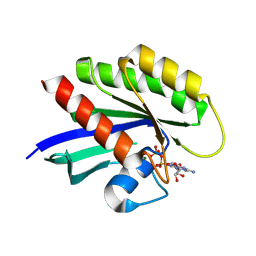 | | Crystal Structure of small molecule acrylamide 16 covalently bound to K-Ras G12C | | Descriptor: | 1-{4-[(2,4-dichlorophenoxy)acetyl]piperazin-1-yl}propan-1-one, GUANOSINE-5'-DIPHOSPHATE, K-Ras GTPase, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
4LPK
 
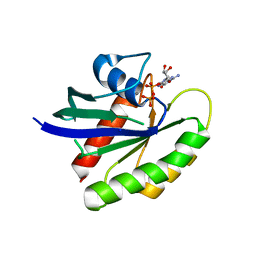 | | Crystal Structure of K-Ras WT, GDP-bound | | Descriptor: | CALCIUM ION, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-07-16 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
4LUC
 
 | | Crystal Structure of small molecule disulfide 6 bound to K-Ras G12C | | Descriptor: | CALCIUM ION, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-07-25 | | Release date: | 2013-11-27 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
