4MJS
 
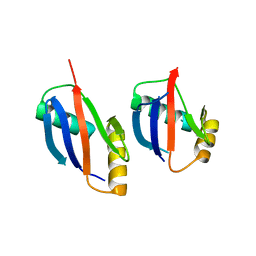 | | crystal structure of a PB1 complex | | Descriptor: | 1,2-ETHANEDIOL, Protein kinase C zeta type, Sequestosome-1 | | Authors: | Ren, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2013-09-04 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical insights into the homotypic PB1-PB1 complex between PKC zeta and p62
Sci China Life Sci, 57, 2014
|
|
5GKK
 
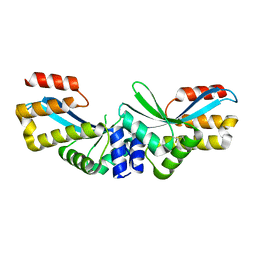 | |
4UWX
 
 | | Structure of liprin-alpha3 in complex with mDia1 Diaphanous- inhibitory domain | | Descriptor: | LIPRIN-ALPHA-3, NICKEL (II) ION, PROTEIN DIAPHANOUS HOMOLOG 1, ... | | Authors: | Brenig, J, de Boor, S, Knyphausen, P, Kuhlmann, N, Wroblowski, S, Baldus, L, Scislowski, L, Artz, O, Trauschies, P, Baumann, U, Neundorf, I, Lammers, M. | | Deposit date: | 2014-08-15 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Basis for the Inhibitory Effect of Liprin-Alpha3 on Mouse Diaphanous 1 (Mdia1) Function.
J.Biol.Chem., 290, 2015
|
|
1REV
 
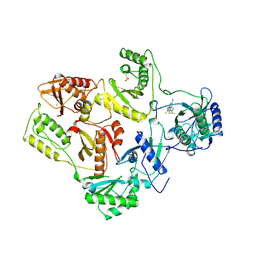 | | HIV-1 REVERSE TRANSCRIPTASE | | Descriptor: | 4-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, HIV-1 REVERSE TRANSCRIPTASE, MAGNESIUM ION | | Authors: | Ren, J, Esnouf, R, Hopkins, A, Ross, C, Jones, Y, Stammers, D, Stuart, D. | | Deposit date: | 1995-09-17 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of HIV-1 reverse transcriptase complexed with 9-chloro-TIBO: lessons for inhibitor design.
Structure, 3, 1995
|
|
1RTH
 
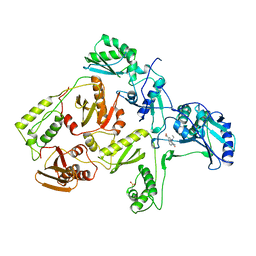 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | 6,11-DIHYDRO-11-ETHYL-6-METHYL-9-NITRO-5H-PYRIDO[2,3-B][1,5]BENZODIAZEPIN-5-ONE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-05-03 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
1RTI
 
 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | 1-(2-HYDROXYETHYLOXYMETHYL)-6-PHENYL THIOTHYMINE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-05-03 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
3KJK
 
 | | Crystal structure of NMB1025, a member of YjgF protein family, from Neisseria meningitidis (monoclinic crystal form) | | Descriptor: | NMB1025 protein | | Authors: | Ren, J, Sainsbury, S, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-11-03 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of NMB1025, a member of YjgF protein family, from Neisseria meningitidis
To be Published
|
|
3KJJ
 
 | | Crystal structure of NMB1025, a member of YjgF protein family, from Neisseria meningitidis (hexagonal crystal form) | | Descriptor: | GLYCEROL, NMB1025 protein | | Authors: | Ren, J, Sainsbury, S, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-11-03 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of NMB1025, a member of YjgF protein family, from Neisseria meningitidis
To be Published
|
|
1MU2
 
 | | CRYSTAL STRUCTURE OF HIV-2 REVERSE TRANSCRIPTASE | | Descriptor: | GLYCEROL, HIV-2 RT, SULFATE ION | | Authors: | Ren, J, Bird, L.E, Chamberlain, P.P, Stewart-Jones, G.B, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-09-23 | | Release date: | 2002-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of HIV-2 reverse transcriptase at 2.35-A resolution and the mechanism of resistance to non-nucleoside inhibitors
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3KXA
 
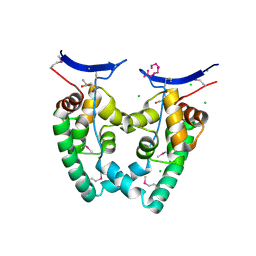 | | Crystal Structure of NGO0477 from Neisseria gonorrhoeae | | Descriptor: | ASPARAGINE, CHLORIDE ION, Putative uncharacterized protein, ... | | Authors: | Ren, J, Sainsbury, S, Nettleship, J.E, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-12-02 | | Release date: | 2010-01-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of NGO0477 from Neisseria gonorrhoeae reveals a novel protein fold incorporating a helix-turn-helix motif.
Proteins, 78, 2010
|
|
3CAM
 
 | |
3DLG
 
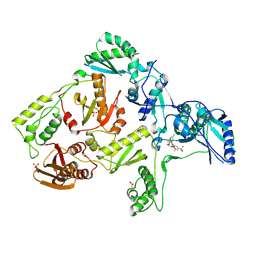 | | Crystal structure of hiv-1 reverse transcriptase in complex with GW564511. | | Descriptor: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, P51 RT, PHOSPHATE ION, ... | | Authors: | Ren, J, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2008-06-27 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
3DMJ
 
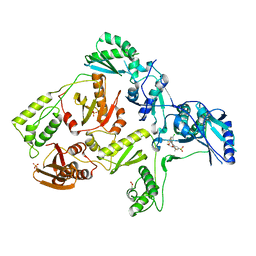 | | CRYSTAL STRUCTURE OF HIV-1 V106A and Y181C MUTANT REVERSE TRANSCRIPTASE IN COMPLEX WITH GW564511. | | Descriptor: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2008-07-01 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
3DLE
 
 | |
3DM2
 
 | | Crystal structure of HIV-1 K103N mutant reverse transcriptase in complex with GW564511. | | Descriptor: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
2A0J
 
 | | Crystal Structure of Nitrogen Regulatory Protein IIA-Ntr from Neisseria meningitidis | | Descriptor: | PTS system, nitrogen regulatory IIA protein | | Authors: | Ren, J, Sainsbury, S, Berrow, N.S, Alderton, D, Nettleship, J.E, Stammers, D.K, Saunders, N.J, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2005-06-16 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of nitrogen regulatory protein IIANtr from Neisseria meningitidis
Bmc Struct.Biol., 5, 2005
|
|
1HML
 
 | | ALPHA_LACTALBUMIN POSSESSES A DISTINCT ZINC BINDING SITE | | Descriptor: | ALPHA-LACTALBUMIN, CALCIUM ION, SULFATE ION, ... | | Authors: | Ren, J, Stuart, D.I, Acharya, K.R. | | Deposit date: | 1994-09-29 | | Release date: | 1995-01-26 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Alpha-lactalbumin possesses a distinct zinc binding site.
J.Biol.Chem., 268, 1993
|
|
1AHC
 
 | |
1AHA
 
 | | THE N-GLYCOSIDASE MECHANISM OF RIBOSOME-INACTIVATING PROTEINS IMPLIED BY CRYSTAL STRUCTURES OF ALPHA-MOMORCHARIN | | Descriptor: | ADENINE, ALPHA-MOMORCHARIN | | Authors: | Ren, J, Wang, Y, Dong, Y, Stuart, D.I. | | Deposit date: | 1994-01-07 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The N-glycosidase mechanism of ribosome-inactivating proteins implied by crystal structures of alpha-momorcharin.
Structure, 2, 1994
|
|
1AHB
 
 | | THE N-GLYCOSIDASE MECHANISM OF RIBOSOME-INACTIVATING PROTEINS IMPLIED BY CRYSTAL STRUCTURES OF ALPHA-MOMORCHARIN | | Descriptor: | ALPHA-MOMORCHARIN, FORMYCIN-5'-MONOPHOSPHATE | | Authors: | Ren, J, Wang, Y, Dong, Y, Stuart, D.I. | | Deposit date: | 1994-01-07 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The N-glycosidase mechanism of ribosome-inactivating proteins implied by crystal structures of alpha-momorcharin.
Structure, 2, 1994
|
|
2BZ0
 
 | | Crystal Structure of E. coli GTP cyclohydrolase II in complex with GTP analogue, GMPcPP, and Zinc | | Descriptor: | GTP CYCLOHYDROLASE II, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Ren, J, Kotaka, M, Lockyer, M, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2005-08-09 | | Release date: | 2005-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | GTP Cyclohydrolase II Structure and Mechanism.
J.Biol.Chem., 280, 2005
|
|
2BZ1
 
 | | CRYSTAL STRUCTURE OF APO E. COLI GTP CYCLOHYDROLASE II | | Descriptor: | 2-AMINOETHANESULFONIC ACID, GLYCEROL, GTP CYCLOHYDROLASE II, ... | | Authors: | Ren, J, Kotaka, M, Lockyer, M, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2005-08-09 | | Release date: | 2005-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | GTP Cyclohydrolase II Structure and Mechanism.
J.Biol.Chem., 280, 2005
|
|
2HNY
 
 | | Crystal Structure of E138K Mutant HIV-1 Reverse Transcriptase in Complex with Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-13 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
2HNZ
 
 | | Crystal Structure of E138K Mutant HIV-1 Reverse Transcriptase in Complex with PETT-2 | | Descriptor: | 1-[2-(4-ETHOXY-3-FLUOROPYRIDIN-2-YL)ETHYL]-3-(5-METHYLPYRIDIN-2-YL)THIOUREA, PHOSPHATE ION, Reverse transcriptase/ribonuclease H | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-13 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
2HND
 
 | | Crystal Structure of K101E Mutant HIV-1 Reverse Transcriptase in Complex with Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-12 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
