7ZRL
 
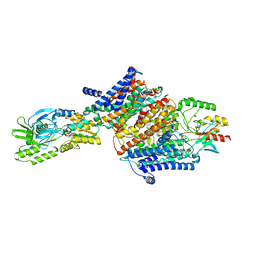 | | Cryo-EM map of the unphosphorylated KdpFABC complex in the E2-P conformation, under turnover conditions | | Descriptor: | POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, Potassium-transporting ATPase KdpC subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRK
 
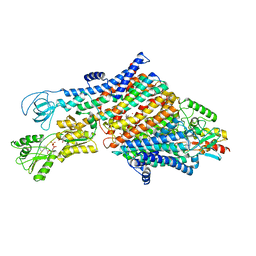 | | Cryo-EM map of the WT KdpFABC complex in the E1-P_ADP conformation, under turnover conditions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARDIOLIPIN, POTASSIUM ION, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRG
 
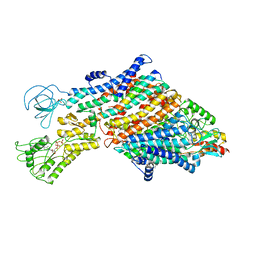 | | Cryo-EM map of the WT KdpFABC complex in the E1_ATPearly conformation, under turnover conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARDIOLIPIN, POTASSIUM ION, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Rheinberger, J, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRH
 
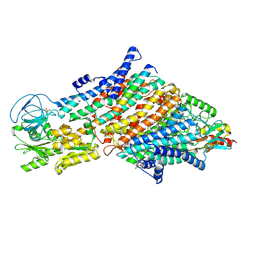 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRI
 
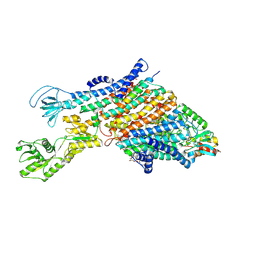 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRJ
 
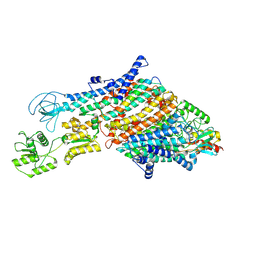 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
6QMB
 
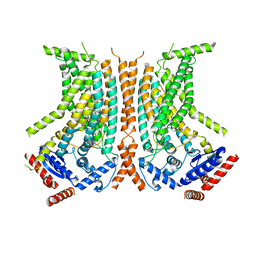 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in nanodisc (closed state) | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QM9
 
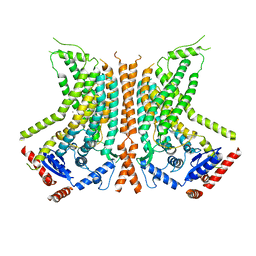 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in nanodisc (open state) | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QM4
 
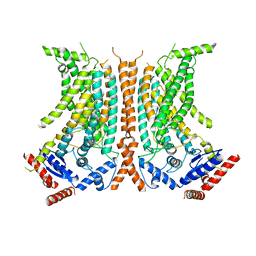 | | Cryo-EM structure of calcium-free nhTMEM16 lipid scramblase in nanodisc | | Descriptor: | Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QM6
 
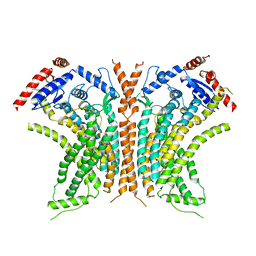 | | Cryo-EM structure of calcium-free nhTMEM16 lipid scramblase in DDM | | Descriptor: | Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QMA
 
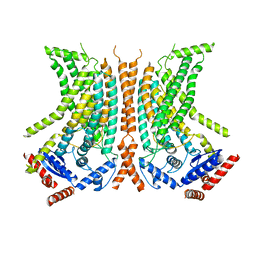 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in nanodisc (intermediate state) | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QM5
 
 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in DDM | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
7B5E
 
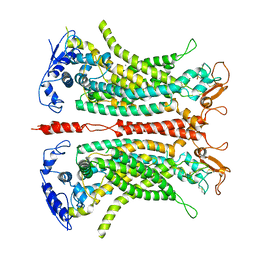 | | Structure of calcium-bound mTMEM16A(ac)-I551A chloride channel at 4.1 A resolution | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Lam, A.K.M, Rheinberger, J, Paulino, C, Dutzler, R. | | Deposit date: | 2020-12-03 | | Release date: | 2021-02-10 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Gating the pore of the calcium-activated chloride channel TMEM16A.
Nat Commun, 12, 2021
|
|
7B5D
 
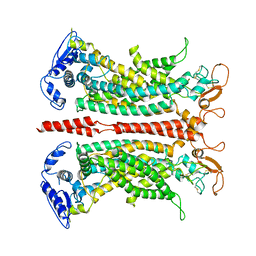 | |
7B5C
 
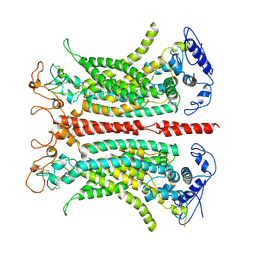 | | Structure of calcium-bound mTMEM16A(ac) chloride channel at 3.7 A resolution | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Lam, A.K.M, Rheinberger, J, Paulino, C, Dutzler, R. | | Deposit date: | 2020-12-03 | | Release date: | 2021-02-10 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Gating the pore of the calcium-activated chloride channel TMEM16A.
Nat Commun, 12, 2021
|
|
7AHC
 
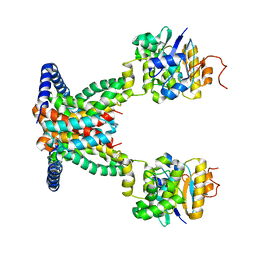 | | OpuA apo inward-facing | | Descriptor: | ABC transporter permease subunit, ABC-type proline/glycine betaine transport system ATPase component | | Authors: | Sikkema, H.R, Rheinberger, J, Paulino, C, Poolman, B. | | Deposit date: | 2020-09-24 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Gating by ionic strength and safety check by cyclic-di-AMP in the ABC transporter OpuA.
Sci Adv, 6, 2020
|
|
7AHD
 
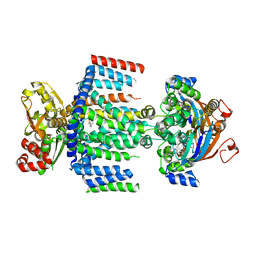 | | OpuA (E190Q) occluded | | Descriptor: | ABC transporter permease subunit, ABC-type proline/glycine betaine transport system ATPase component, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Sikkema, H.R, Rheinberger, J, Paulino, C, Poolman, B. | | Deposit date: | 2020-09-24 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Gating by ionic strength and safety check by cyclic-di-AMP in the ABC transporter OpuA.
Sci Adv, 6, 2020
|
|
7AHH
 
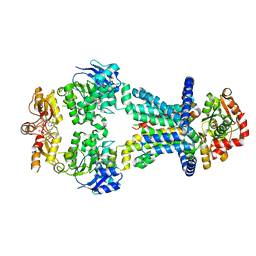 | | OpuA inhibited inward-facing, SBD docked | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ABC transporter permease subunit, ABC-type proline/glycine betaine transport system ATPase component, ... | | Authors: | Sikkema, H.R, Rheinberger, J, Paulino, C, Poolman, B. | | Deposit date: | 2020-09-24 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Gating by ionic strength and safety check by cyclic-di-AMP in the ABC transporter OpuA.
Sci Adv, 6, 2020
|
|
7AHE
 
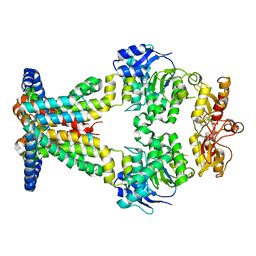 | | OpuA inhibited inward facing | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ABC transporter permease subunit, ABC-type proline/glycine betaine transport system ATPase component | | Authors: | Sikkema, H.R, Rheinberger, J, Paulino, C, Poolman, B. | | Deposit date: | 2020-09-24 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Gating by ionic strength and safety check by cyclic-di-AMP in the ABC transporter OpuA.
Sci Adv, 6, 2020
|
|
8B8G
 
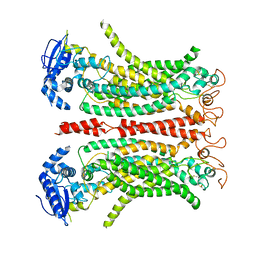 | | Cryo-EM structure of Ca2+-free mTMEM16F F518H mutant in Digitonin | | Descriptor: | Anoctamin-6 | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8K
 
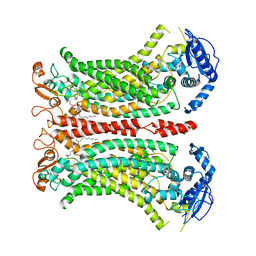 | | Cryo-EM structure of Ca2+-bound mTMEM16F N562A mutant in Digitonin closed/closed | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8J
 
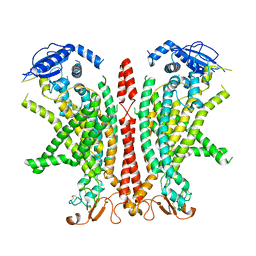 | | Cryo-EM structure of Ca2+-bound mTMEM16F F518H mutant in Digitonin | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8BC0
 
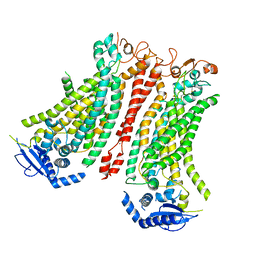 | | Cryo-EM structure of Ca2+-bound mTMEM16F F518A Q623A mutant in GDN open/closed | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8BC1
 
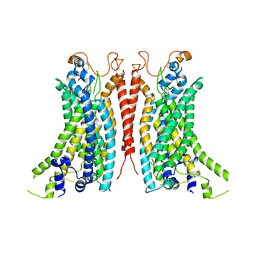 | | Cryo-EM Structure of Ca2+-bound mTMEM16F F518A_Q623A mutant in GDN | | Descriptor: | Anoctamin-6,mTMEM16F, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8M
 
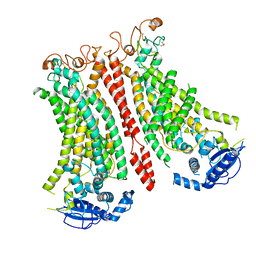 | | Cryo-EM structure of Ca2+-bound mTMEM16F N562A mutant in Digitonin open/closed | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
