6USD
 
 | | Barrier-to-autointegration factor soaked in ethanol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-25 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6URZ
 
 | | Barrier-to-autointegration factor soaked in methanol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL, METHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7ULK
 
 | | Human TRAP1 NM in complex with 42C | | Descriptor: | CALCIUM ION, Heat shock protein 75 kDa, mitochondrial, ... | | Authors: | Stachowski, T.R, Nithianantham, S, Vanarotti, M, Fischer, M. | | Deposit date: | 2022-04-05 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Pan-HSP90 ligand binding reveals isoform-specific differences in plasticity and water networks.
Protein Sci., 32, 2023
|
|
7ULJ
 
 | | Hsp90b N-terminal domain in complex with 42C | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Heat shock protein HSP 90-beta, ... | | Authors: | Stachowski, T.R, Nithianantham, S, Vanarotti, M, Fischer, M. | | Deposit date: | 2022-04-05 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Pan-HSP90 ligand binding reveals isoform-specific differences in plasticity and water networks.
Protein Sci., 32, 2023
|
|
7ULL
 
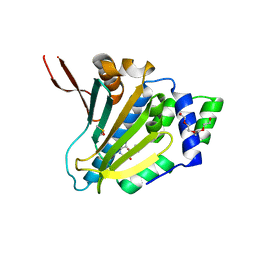 | | Human Grp94 N-terminal domain in complex with 42C | | Descriptor: | DIMETHYL SULFOXIDE, Endoplasmin, GLYCEROL, ... | | Authors: | Stachowski, T.R, Nithianantham, S, Vanarotti, M, Fischer, M. | | Deposit date: | 2022-04-05 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Pan-HSP90 ligand binding reveals isoform-specific differences in plasticity and water networks.
Protein Sci., 32, 2023
|
|
6URJ
 
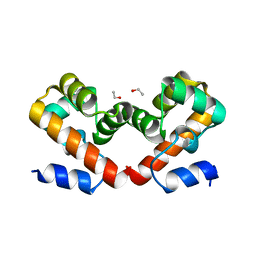 | | Barrier-to-autointegration factor soaked in Acetone: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6USB
 
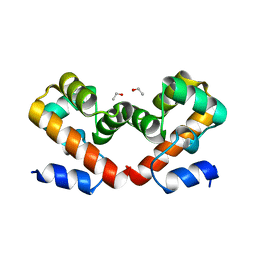 | | Barrier-to-autointegration factor soaked in urea: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-25 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6US1
 
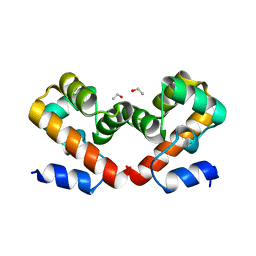 | | Barrier-to-autointegration factor soaked in isobutanol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6US7
 
 | | Barrier-to-autointegration factor soaked in TMAO: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6URN
 
 | | Barrier-to-autointegration factor t-butanol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6US0
 
 | | Barrier-to-autointegration factor soaked in R,S,R-bisfuranol (RSR): 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6NBS
 
 | | WT ERK2 with compound 2507-8 | | Descriptor: | (5S)-5-benzyl-4,5-dihydro-1H-imidazol-2-amine, GLYCEROL, Mitogen-activated protein kinase 1, ... | | Authors: | Sammons, R.M, Perry, N.A, Cho, E.J, Kaoud, T.S, Zamora-Olivares, D.P, Piserchio, A, Houghten, R.A, Giulianotti, M, Li, Y, Debevec, G, Gurevich, V.V, Ghose, R, Iverson, T.M, Dalby, K.N. | | Deposit date: | 2018-12-10 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Class of Common Docking Domain Inhibitors That Prevent ERK2 Activation and Substrate Phosphorylation.
Acs Chem.Biol., 14, 2019
|
|
7Q21
 
 | | III2-IV2 respiratory supercomplex from Corynebacterium glutamicum | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Kovalova, T, Moe, A, Krol, S, Yanofsky, D.J, Bott, M, Sjostrand, D, Rubinstein, J.L, Hogbom, M, Brzezinski, P. | | Deposit date: | 2021-10-22 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The respiratory supercomplex from C. glutamicum.
Structure, 30, 2022
|
|
4QCJ
 
 | | Crystal Structure of OdhI from Corynebacterium glutamicum | | Descriptor: | Oxoglutarate dehydrogenase inhibitor | | Authors: | Labahn, J, Raasch, K, Eggeling, L, Bocola, M, Leitner, A, Bott, M. | | Deposit date: | 2014-05-12 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interaction of 2-oxoglutarate dehydrogenase OdhA with its inhibitor OdhI in Corynebacterium glutamicum: Mutants and a model.
J.Biotechnol., 191, 2014
|
|
3PCQ
 
 | | Femtosecond X-ray protein Nanocrystallography | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Chapman, H.N, Fromme, P, Barty, A, White, T.A, Kirian, R.A, Aquila, A, Hunter, M.S, Schulz, J, Deponte, D.P, Weierstall, U, Doak, R.B, Maia, F.R.N.C, Martin, A.V, Schlichting, I, Lomb, L, Coppola, N, Shoeman, R.L, Epp, S.W, Hartmann, R, Rolles, D, Rudenko, A, Foucar, L, Kimmel, N, Weidenspointner, G, Holl, P, Liang, M, Barthelmess, M, Caleman, C, Boutet, S, Bogan, M.J, Krzywinski, J, Bostedt, C, Bajt, S, Gumprecht, L, Rudek, B, Erk, B, Schmidt, C, Homke, A, Reich, C, Pietschner, D, Struder, L, Hauser, G, Gorke, H, Ullrich, J, Herrmann, S, Schaller, G, Schopper, F, Soltau, H, Kuhnel, K.-U, Messerschmidt, M, Bozek, J.D, Hau-Riege, S.P, Frank, M, Hampton, C.Y, Sierra, R, Starodub, D, Williams, G.J, Hajdu, J, Timneanu, N, Seibert, M.M, Andreasson, J, Rocker, A, Jonsson, O, Svenda, M, Stern, S, Nass, K, Andritschke, R, Schroter, C.-D, Krasniqi, F, Bott, M, Schmidt, K.E, Wang, X, Grotjohann, I, Holton, J.M, Barends, T.R.M, Neutze, R, Marchesini, S, Fromme, R, Schorb, S, Rupp, D, Adolph, M, Gorkhover, T, Andersson, I, Hirsemann, H, Potdevin, G, Graafsma, H, Nilsson, B, Spence, J.C.H. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (8.984 Å) | | Cite: | Femtosecond X-ray protein nanocrystallography.
Nature, 470, 2011
|
|
4NAL
 
 | | Arabidopsis thaliana IspD in complex with tribromodichloro-pseudilin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,4-DICHLORO-6-(3,4,5-TRIBROMO-1H-PYRROL-2-YL)PHENOL, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, ... | | Authors: | Kunfermann, A, Witschel, M, Illarionov, B, Martin, R, Rottmann, M, Hoffken, H.W, Seet, M, Eisenreich, W, Knolker, H.-J, Fischer, M, Bacher, A, Groll, M, Diederich, F. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pseudilins: Halogenated, Allosteric Inhibitors of the Non-Mevalonate Pathway Enzyme IspD.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NAI
 
 | | Arabidopsis thaliana IspD apo | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, CADMIUM ION, ... | | Authors: | Kunfermann, A, Witschel, M, Illarionov, B, Martin, R, Rottmann, M, Hoffken, H.W, Seet, M, Eisenreich, W, Knolker, H.-J, Fischer, M, Bacher, A, Groll, M, Diederich, F. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pseudilins: Halogenated, Allosteric Inhibitors of the Non-Mevalonate Pathway Enzyme IspD.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NAK
 
 | | Arabidopsis thaliana IspD in complex with pentabromo-pseudilin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, ... | | Authors: | Kunfermann, A, Witschel, M, Illarionov, B, Martin, R, Rottmann, M, Hoffken, H.W, Seet, M, Eisenreich, W, Knolker, H.-J, Fischer, M, Bacher, A, Groll, M, Diederich, F. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pseudilins: Halogenated, Allosteric Inhibitors of the Non-Mevalonate Pathway Enzyme IspD.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NAN
 
 | | Arabidopsis thaliana IspD in complex with tetrabromo-pseudilin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, ... | | Authors: | Kunfermann, A, Witschel, M, Illarionov, B, Martin, R, Rottmann, M, Hoffken, H.W, Seet, M, Eisenreich, W, Knolker, H.-J, Fischer, M, Bacher, A, Groll, M, Diederich, F. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pseudilins: Halogenated, Allosteric Inhibitors of the Non-Mevalonate Pathway Enzyme IspD.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5L8S
 
 | | The crystal structure of a cold-adapted acylaminoacyl peptidase reveals a novel quaternary architecture based on the arm-exchange mechanism | | Descriptor: | Amino acyl peptidase, SULFATE ION | | Authors: | Brocca, S, Ferrari, C, Barbiroli, A, Pesce, A, Lotti, M, Nardini, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A bacterial acyl aminoacyl peptidase couples flexibility and stability as a result of cold adaptation.
FEBS J., 283, 2016
|
|
3ISN
 
 | | Crystal structure of HIV-1 RT bound to A 6-vinylpyrimidine inhibitor | | Descriptor: | 6-ethenyl-N,N-dimethyl-2-(methylsulfonyl)pyrimidin-4-amine, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Ennifar, E, Freisz, S, Bec, G, Dumas, P, Botta, M, Radi, M. | | Deposit date: | 2009-08-26 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of HIV-1 Reverse Transcriptase Bound to a Non-Nucleoside Inhibitor with a Novel Mechanism of Action
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3ITH
 
 | | Crystal structure of the HIV-1 reverse transcriptase bound to a 6-vinylpyrimidine inhibitor | | Descriptor: | 6-ethenyl-N,N-dimethyl-2-(methylsulfonyl)pyrimidin-4-amine, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Freisz, S, Bec, G, Wolff, P, Dumas, P, Radi, M, Botta, M. | | Deposit date: | 2009-08-28 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of HIV-1 Reverse Transcriptase Bound to a Non-Nucleoside Inhibitor with a Novel Mechanism of Action
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
7A04
 
 | | Structure of human CKa1 in complex with compound b | | Descriptor: | 1,2-ETHANEDIOL, 1-(phenylmethyl)-4-pyrrolidin-1-yl-pyridin-1-ium, Choline kinase alpha | | Authors: | Serran-Aguilera, L, Mariotto, E, Rubbini, G, Castro Navas, F.C, Marco, C, Carrasco-Jimenez, M.P, Ballarotto, M, Macchiarulo, A, Hurtado-Guerrero, R, Viola, G, Lopez-Cara, L.C. | | Deposit date: | 2020-08-06 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Synthesis, biological evaluation, in silico modeling and crystallization of novel small monocationic molecules with potent antiproliferative activity by dual mechanism.
Eur.J.Med.Chem., 207, 2020
|
|
7A06
 
 | | Structure of human CKa1 in complex with compound o | | Descriptor: | 1,2-ETHANEDIOL, 1-[(4-phenylphenyl)methyl]-4-pyrrolidin-1-yl-pyridine, Choline kinase alpha | | Authors: | Serran-Aguilera, L, Mariotto, E, Rubbini, G, Castro Navas, F.C, Marco, C, Carrasco-Jimenez, M.P, Ballarotto, M, Macchiarulo, A, Hurtado-Guerrero, R, Viola, G, Lopez-Cara, L.C. | | Deposit date: | 2020-08-06 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis, biological evaluation, in silico modeling and crystallization of novel small monocationic molecules with potent antiproliferative activity by dual mechanism.
Eur.J.Med.Chem., 207, 2020
|
|
2BAG
 
 | | 3D Structure of Torpedo californica acetylcholinesterase complexed with Ganstigmine | | Descriptor: | 1S,3AS,8AS-TRIMETHYL-1-OXIDO-1,2,3,3A,8,8A-HEXAHYDROPYRROLO[2,3-B]INDOL-5-YL 2-ETHYLPHENYLCARBAMATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lamba, D, Bartolucci, C, Siotto, M, Racchi, M, Villetti, G, Delcanale, M. | | Deposit date: | 2005-10-14 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Determinants of Torpedo californica Acetylcholinesterase Inhibition by the Novel and Orally Active Carbamate Based Anti-Alzheimer Drug Ganstigmine (CHF-2819)
J.Med.Chem., 49, 2006
|
|
