5J7C
 
 | |
5JQK
 
 | |
5JR6
 
 | |
5J7K
 
 | |
5KYO
 
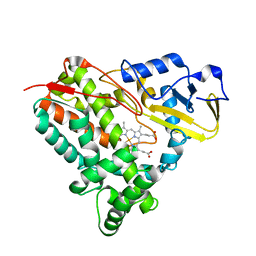 | | Crystal Structure of CYP101J2 | | Descriptor: | CYP101J2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Unterweger, B, Drinkwater, N, Dumsday, G.J, McGowan, S. | | Deposit date: | 2016-07-22 | | Release date: | 2017-01-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of cytochrome P450 monooxygenase CYP101J2 from Sphingobium yanoikuyae strain B2.
Proteins, 85, 2017
|
|
8T6H
 
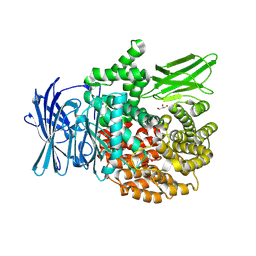 | |
8T83
 
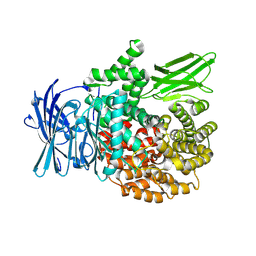 | |
8T7P
 
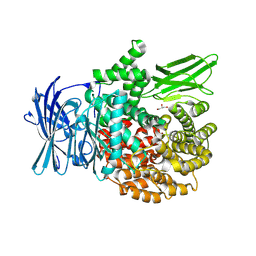 | | X-ray crystal structure of PfA-M1(M462S) | | Descriptor: | Aminopeptidase N, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Yang, W, Drinkwater, N, Webb, C.T, McGowan, S. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational dynamics of the Plasmodium falciparum M1 aminopeptidase.
To Be Published
|
|
6NVB
 
 | | Crystal structure of the inhibitor-free form of the serine protease kallikrein-4 | | Descriptor: | GLYCEROL, Kallikrein-4, SULFATE ION | | Authors: | Riley, B.T, Buckle, A.M, McGowan, S. | | Deposit date: | 2019-02-04 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.636 Å) | | Cite: | Crystal structure of the inhibitor-free form of the serine protease kallikrein-4.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7RIE
 
 | | Plasmodium falciparum M17 in complex with inhibitor MIPS2571 | | Descriptor: | CARBONATE ION, M17 leucyl aminopeptidase, N-{(1R)-2-(hydroxyamino)-1-[4'-(hydroxymethyl)[1,1'-biphenyl]-4-yl]-2-oxoethyl}-2,2-dimethylpropanamide, ... | | Authors: | Webb, C.T, McGowan, S. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Genetic and chemical validation of Plasmodium falciparum aminopeptidase Pf A-M17 as a drug target in the hemoglobin digestion pathway.
Elife, 11, 2022
|
|
5CBM
 
 | | Crystal structure of PfA-M17 with virtual ligand inhibitor | | Descriptor: | (2S)-2-{[(R)-[(R)-amino(phenyl)methyl](hydroxy)phosphoryl]methyl}-4-methylpentanoic acid, CARBONATE ION, GLYCEROL, ... | | Authors: | Ruggeri, C, Drinkwater, N, McGowan, S. | | Deposit date: | 2015-07-01 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and Validation of a Potent Dual Inhibitor of the P. falciparum M1 and M17 Aminopeptidases Using Virtual Screening.
Plos One, 10, 2015
|
|
6EE3
 
 | | X-ray crystal structure of Pf-M1 in complex with inhibitor (6k) and catalytic zinc ion | | Descriptor: | (1R,2r,3S,5R,7R)-N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)ethyl]tricyclo[3.3.1.1~3,7~]decane-2-carboxamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2018-08-13 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Hydroxamic Acid Inhibitors Provide Cross-Species Inhibition of Plasmodium M1 and M17 Aminopeptidases.
J. Med. Chem., 62, 2019
|
|
6EED
 
 | | X-ray crystal structure of Pf-M1 in complex with inhibitor (6p) and catalytic zinc ion | | Descriptor: | (2R)-2-[(cyclohexylacetyl)amino]-N-hydroxy-2-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)acetamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2018-08-13 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hydroxamic Acid Inhibitors Provide Cross-Species Inhibition of Plasmodium M1 and M17 Aminopeptidases.
J. Med. Chem., 62, 2019
|
|
6EA2
 
 | |
6EAB
 
 | |
6EAA
 
 | |
6EE4
 
 | | X-ray crystal structure of Pf-M1 in complex with inhibitor (6m) and catalytic zinc ion | | Descriptor: | (2R)-2-[(cyclopropylacetyl)amino]-N-hydroxy-2-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)acetamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2018-08-13 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Hydroxamic Acid Inhibitors Provide Cross-Species Inhibition of Plasmodium M1 and M17 Aminopeptidases.
J. Med. Chem., 62, 2019
|
|
6EE2
 
 | |
6EE6
 
 | | X-ray crystal structure of Pf-M1 in complex with inhibitor (6o) and catalytic zinc ion | | Descriptor: | (2R)-2-[(cyclopentylacetyl)amino]-N-hydroxy-2-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)acetamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2018-08-13 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hydroxamic Acid Inhibitors Provide Cross-Species Inhibition of Plasmodium M1 and M17 Aminopeptidases.
J. Med. Chem., 62, 2019
|
|
7KWW
 
 | | X-ray Crystal Structure of PlyCB Mutant K59H | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | Authors: | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-12-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
7KWY
 
 | | X-ray Crystal Structure of PlyCB Mutant R66K | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | Authors: | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-12-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
7KWT
 
 | | X-ray Crystal Structure of PlyCB Mutant Y28H | | Descriptor: | PlyCB | | Authors: | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-12-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
7K5K
 
 | | Plasmodium vivax M17 leucyl aminopeptidase Pv-M17 | | Descriptor: | CARBONATE ION, M17 leucyl aminopeptidase, putative, ... | | Authors: | Malcolm, T.R, McGowan, S, Belousoff, M.J. | | Deposit date: | 2020-09-17 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Active site metals mediate an oligomeric equilibrium in Plasmodium M17 aminopeptidases.
J.Biol.Chem., 296, 2020
|
|
6EEE
 
 | | X-ray crystal structure of Pf-M17 in complex with inhibitor (6k) and regulatory zinc ion | | Descriptor: | (1R,2r,3S,5R,7R)-N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)ethyl]tricyclo[3.3.1.1~3,7~]decane-2-carboxamide, 1,2-ETHANEDIOL, CARBONATE ION, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2018-08-13 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Hydroxamic Acid Inhibitors Provide Cross-Species Inhibition of Plasmodium M1 and M17 Aminopeptidases.
J. Med. Chem., 62, 2019
|
|
6EA1
 
 | | X-ray crystal structure of Pf-M1 in complex with inhibitor (6da) and catalytic zinc ion | | Descriptor: | (2R)-2,3,3,3-tetrafluoro-N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)ethyl]propanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2018-08-02 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.815 Å) | | Cite: | Hydroxamic Acid Inhibitors Provide Cross-Species Inhibition of Plasmodium M1 and M17 Aminopeptidases.
J. Med. Chem., 62, 2019
|
|
