3U1L
 
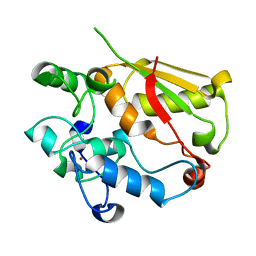 | | Structure of the mRNA splicing complex component Cwc2 | | Descriptor: | Pre-mRNA-splicing factor CWC2, ZINC ION | | Authors: | Lu, P, Lu, G, Yan, C, Wang, L, Li, W, Yin, P. | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of the mRNA splicing complex component Cwc2: insights into RNA recognition
Biochem.J., 441, 2012
|
|
4NFB
 
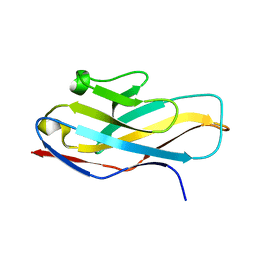 | | Structure of paired immunoglobulin-like type 2 receptor (PILR ) | | Descriptor: | Paired immunoglobulin-like type 2 receptor alpha | | Authors: | Lu, Q, Lu, G, Qi, J, Li, Y, Zhang, Y, Wang, H, Fan, Z, Yan, J, Gao, G. | | Deposit date: | 2013-10-31 | | Release date: | 2014-05-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PILR alpha and PILR beta have a siglec fold and provide the basis of binding to sialic acid
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4EWJ
 
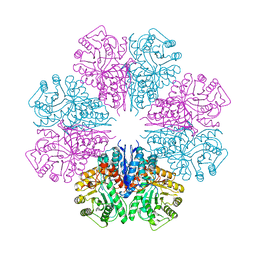 | | structure of the enloase from Streptococcus suis serotype 2 | | Descriptor: | Enolase 2 | | Authors: | Lu, Q, Lu, H, Qi, J, Lu, G, Gao, G.F. | | Deposit date: | 2012-04-27 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | An octamer of enolase from Streptococcus suis.
Protein Cell, 3, 2012
|
|
7XJZ
 
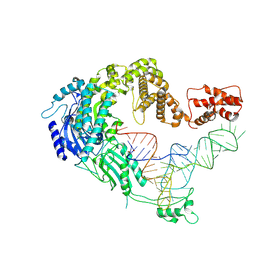 | | Cryo-EM strucrture of Oryza sativa plastid glycyl-tRNA synthetase in complex with tRNA (tRNA binding state) | | Descriptor: | Glycine--tRNA ligase, tRNA(gly) | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
7XJY
 
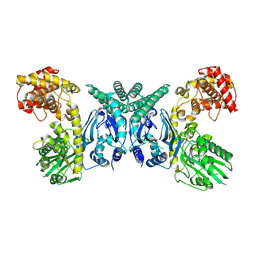 | | Cryo-EM structure of Oryza sativa plastid glycyl-tRNA synthetase (apo form) | | Descriptor: | Glycine--tRNA ligase | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
7XOF
 
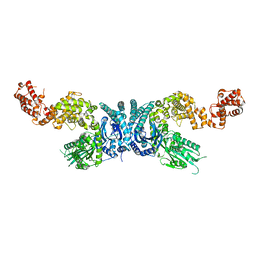 | |
7XK1
 
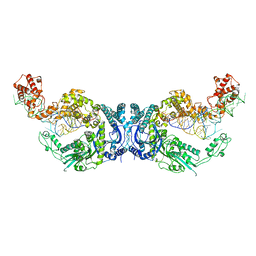 | | Cryo-EM structure of Oryza sativa plastid glycyl-tRNA synthetase in complex with two tRNAs (both in tRNA binding states) | | Descriptor: | Glycine--tRNA ligase, tRNA(gly) | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
7XK0
 
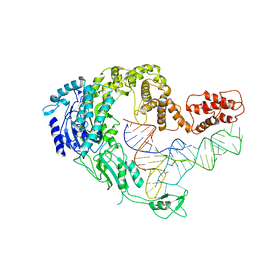 | | Cryo-EM strucrture of Oryza sativa plastid glycyl-tRNA synthetase in complex with tRNA (tRNA locked state) | | Descriptor: | Glycine--tRNA ligase, tRNA(gly) | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
4NFC
 
 | | Structure of paired immunoglobulin-like type 2 receptor (PILR ) | | Descriptor: | Paired immunoglobulin-like type 2 receptor beta | | Authors: | Lu, Q, Lu, G, Qi, J, Li, Y, Zhang, Y, Wang, H, Fan, Z, Yan, J, Gao, G.F. | | Deposit date: | 2013-10-31 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PILR alpha and PILR beta have a siglec fold and provide the basis of binding to sialic acid
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NFD
 
 | | Structure of PILR L108W mutant in complex with sialic acid | | Descriptor: | N-acetyl-alpha-neuraminic acid, Paired immunoglobulin-like type 2 receptor beta | | Authors: | Lu, Q, Lu, G, Qi, J, Li, Y, Zhang, Y, Wang, H, Fan, Z, Yan, J, Gao, G.F. | | Deposit date: | 2013-10-31 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | PILR alpha and PILR beta have a siglec fold and provide the basis of binding to sialic acid
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3FAN
 
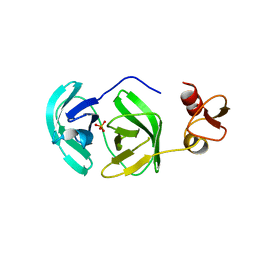 | | Crystal structure of chymotrypsin-like protease/proteinase (3CLSP/Nsp4) of porcine reproductive and respiratory syndrome virus (PRRSV) | | Descriptor: | Non-structural protein, PHOSPHATE ION | | Authors: | Gao, F, Peng, H, Tian, X, Lu, G, Liu, Y, Gao, G.F. | | Deposit date: | 2008-11-17 | | Release date: | 2009-09-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and cleavage specificity of the chymotrypsin-like serine protease (3CLSP/nsp4) of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV).
J.Mol.Biol., 392, 2009
|
|
3FAO
 
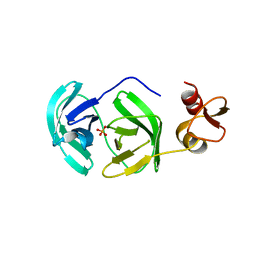 | | Crystal structure of S118A mutant 3CLSP of PRRSV | | Descriptor: | Non-structural protein, PHOSPHATE ION | | Authors: | Gao, F, Peng, H, Tian, X, Lu, G, Liu, Y, Gao, G.F. | | Deposit date: | 2008-11-17 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure and cleavage specificity of the chymotrypsin-like serine protease (3CLSP/nsp4) of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV).
J.Mol.Biol., 392, 2009
|
|
5DO2
 
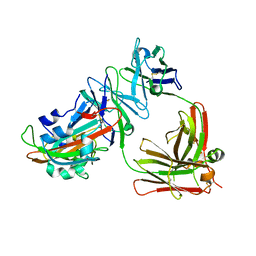 | | Complex structure of MERS-RBD bound with 4C2 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4C2 heavy chain, 4C2 light chain, ... | | Authors: | Li, Y, Wan, Y, Liu, P, Zhao, J, Lu, G, Qi, J, Wang, Q, Lu, X, Wu, Y, Liu, W, Yuen, K.Y, Perlman, S, Gao, G.F, Yan, J. | | Deposit date: | 2015-09-10 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein.
Cell Res., 25, 2015
|
|
2YJY
 
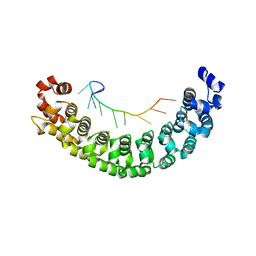 | | A specific and modular binding code for cytosine recognition in PUF domains | | Descriptor: | 5'-R(*AP*UP*UP*GP*CP*AP*UP*AP*UP*AP)-3', PUMILIO HOMOLOG 1 | | Authors: | Dong, S, Wang, Y, Cassidy-Amstutz, C, Lu, G, Qiu, C, Bigler, R, Jezyk, M, Li, C, Hall, T.M.T, Wang, Z. | | Deposit date: | 2011-05-24 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Specific and Modular Binding Code for Cytosine Recognition in Pumilio/Fbf (Puf) RNA-Binding Domains.
J.Biol.Chem., 286, 2011
|
|
3U82
 
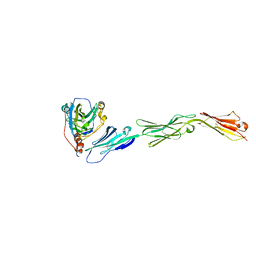 | | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion | | Descriptor: | Envelope glycoprotein D, Poliovirus receptor-related protein 1 | | Authors: | Zhang, N, Yan, J, Lu, G, Guo, Z, Fan, Z, Wang, J, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2011-10-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.164 Å) | | Cite: | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion.
Nat Commun, 2, 2011
|
|
3U83
 
 | | Crystal structure of nectin-1 | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Poliovirus receptor-related protein 1 | | Authors: | Zhang, N, Yan, J, Lu, G, Guo, Z, Fan, Z, Wang, J, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2011-10-15 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion.
Nat Commun, 2, 2011
|
|
2B9K
 
 | |
8BNZ
 
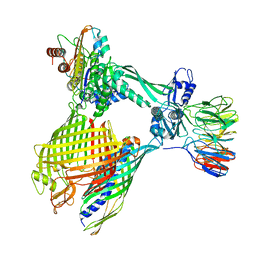 | | BAM-EspP complex structure with BamA-G431C/EspP-N1293C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
8BO2
 
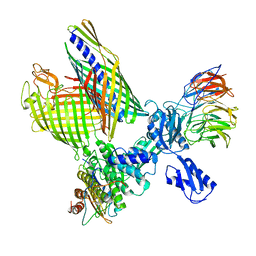 | | BAM-EspP complex structure with BamA-S425C/EspP-S1299C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
4P0C
 
 | | Crystal Structure of NHERF2 PDZ1 Domain in Complex with LPA2 | | Descriptor: | CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF2/Lysophosphatidic acid receptor 2 chimeric protein, THIOCYANATE ION | | Authors: | Holcomb, J, Jiang, Y, Lu, G, Trescott, L, Brunzelle, J, Sirinupong, N, Li, C, Naren, A, Yang, Z. | | Deposit date: | 2014-02-20 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.339 Å) | | Cite: | Structural insights into PDZ-mediated interaction of NHERF2 and LPA2, a cellular event implicated in CFTR channel regulation.
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4MPA
 
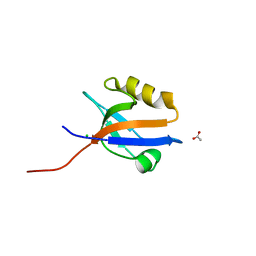 | | Crystal structure of NHERF1-CXCR2 signaling complex in P21 space group | | Descriptor: | ACETIC ACID, CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1, ... | | Authors: | Jiang, Y, Lu, G, Wu, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-09-12 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
4Q3H
 
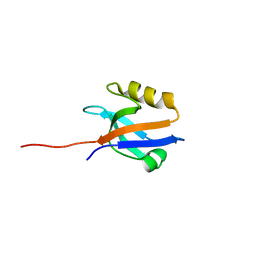 | | The crystal structure of NHERF1 PDZ2 CXCR2 complex revealed by the NHERF1 CXCR2 chimeric protein | | Descriptor: | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Holcomb, J, Jiang, Y, Trescott, L, Lu, G, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2014-04-11 | | Release date: | 2014-05-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.443 Å) | | Cite: | Crystal structure of the NHERF1 PDZ2 domain in complex with the chemokine receptor CXCR2 reveals probable modes of PDZ2 dimerization.
Biochem.Biophys.Res.Commun., 448, 2014
|
|
4NFZ
 
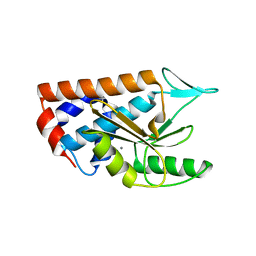 | | Crystal structure of polymerase subunit PA N-terminal endonuclease domain from bat-derived influenza virus H17N10 | | Descriptor: | MANGANESE (II) ION, Polymerase PA | | Authors: | Tefsen, B, Lu, G, Zhu, Y, Haywood, J, Zhao, L, Deng, T, Qi, J, Gao, G.F. | | Deposit date: | 2013-11-01 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The N-Terminal Domain of PA from Bat-Derived Influenza-Like Virus H17N10 Has Endonuclease Activity
J.Virol., 88, 2014
|
|
4GJT
 
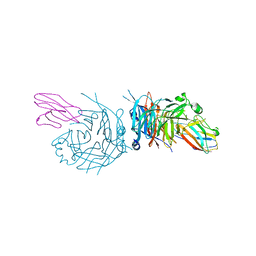 | | complex structure of nectin-4 bound to MV-H | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin glycoprotein, Poliovirus receptor-related protein 4 | | Authors: | Zhang, X, Lu, G, Qi, J, Li, Y, He, Y, Xu, X, Shi, J, Zhang, C, Yan, J, Gao, G.F. | | Deposit date: | 2012-08-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1001 Å) | | Cite: | Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4
Nat.Struct.Mol.Biol., 20, 2013
|
|
5Z38
 
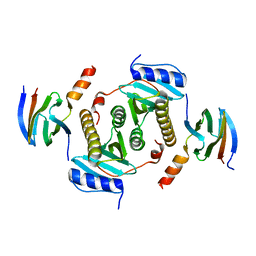 | | Crystal structure of CsrA bound to CesT | | Descriptor: | CesT protein, Truncated-CsrA, wild type CsrA | | Authors: | Ye, F, Yang, F, Yu, R, Lin, X, Qi, J, Chen, Z, Gao, G.F, Lu, G. | | Deposit date: | 2018-01-05 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Molecular basis of binding between the global post-transcriptional regulator CsrA and the T3SS chaperone CesT
Nat Commun, 9, 2018
|
|
