4I3K
 
 | | Crystal structure of a metabolic reductase with 1-hydroxy-6-(4-hydroxybenzyl)-4-methylpyridin-2(1H)-one | | Descriptor: | 1-hydroxy-6-(4-hydroxybenzyl)-4-methylpyridin-2(1H)-one, Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zheng, B, Yao, Y, Liu, Z, Deng, L, Anglin, J.L, Jiang, H, Prasad, B.V.V, Song, Y. | | Deposit date: | 2012-11-26 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3056 Å) | | Cite: | Crystallographic Investigation and Selective Inhibition of Mutant Isocitrate Dehydrogenase.
ACS Med Chem Lett, 4, 2013
|
|
4I3L
 
 | | Crystal structure of a metabolic reductase with 6-benzyl-1-hydroxy-4-methylpyridin-2(1H)-one | | Descriptor: | 6-benzyl-1-hydroxy-4-methylpyridin-2(1H)-one, Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zheng, B, Yao, Y, Liu, Z, Deng, L, Anglin, J.L, Jiang, H, Prasad, B.V.V, Song, Y. | | Deposit date: | 2012-11-26 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.292 Å) | | Cite: | Crystallographic Investigation and Selective Inhibition of Mutant Isocitrate Dehydrogenase.
ACS Med Chem Lett, 4, 2013
|
|
4IMZ
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | Genome polyprotein, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4IN1
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | 3C-like protease, SULFATE ION | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4IMQ
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | 3C-like protease, PEPTIDE INHIBITOR, syc8, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4IN2
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | C-like protease | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4INH
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | DIMETHYL SULFOXIDE, Genome polyprotein, peptide inhibitor, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-04 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
5A55
 
 | | The native structure of GH101 from Streptococcus pneumoniae TIGR4 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDO-ALPHA-N-ACETYLGALACTOSAMINIDASE, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights Into its Mechanism.
J.Biol.Chem., 290, 2015
|
|
5A5A
 
 | | The structure of GH101 E796Q mutant from Streptococcus pneumoniae TIGR4 in complex with PNP-T-antigen | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights Into its Mechanism.
J.Biol.Chem., 290, 2015
|
|
5A57
 
 | | The structure of GH101 from Streptococcus pneumoniae TIGR4 in complex with PUGT | | Descriptor: | (Z)-[(3R,4R,5R,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-ylidene]amino] N-phenylcarbamate, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights Into its Mechanism.
J.Biol.Chem., 290, 2015
|
|
5A58
 
 | | The structure of GH101 D764N mutant from Streptococcus pneumoniae TIGR4 in complex with serinyl T-antigen | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights Into its Mechanism.
J.Biol.Chem., 290, 2015
|
|
5A59
 
 | | The structure of GH101 E796Q mutant from Streptococcus pneumoniae TIGR4 in complex with T-antigen | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights Into its Mechanism.
J.Biol.Chem., 290, 2015
|
|
5A56
 
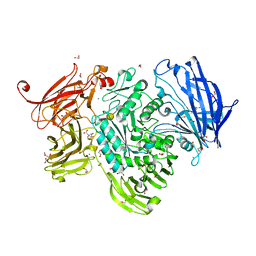 | | The structure of GH101 from Streptococcus pneumoniae TIGR4 in complex with 1-O-methyl-T-antigen | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights into Its Mechanism.
J.Biol.Chem., 290, 2015
|
|
7VUX
 
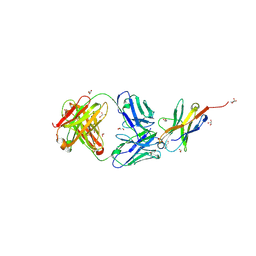 | | Complex structure of PD1 and 609A-Fab | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Huang, H, Zhu, Z, Zhao, J, Jiang, L, Yang, H, Deng, L, Meng, X, Ding, J, Yang, S, Zhao, L, Xu, W, Wang, X. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A strategy for the efficient construction of anti-PD1-based bispecific antibodies with desired IgG-like properties.
Mabs, 14, 2022
|
|
3CAD
 
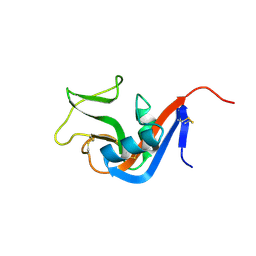 | | Crystal structure of Natural Killer Cell Receptor, Ly49G | | Descriptor: | Lectin-related NK cell receptor LY49G1 | | Authors: | Cho, S. | | Deposit date: | 2008-02-19 | | Release date: | 2008-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Architecture of the Major Histocompatibility Complex Class I-binding Site of Ly49 Natural Killer Cell Receptors.
J.Biol.Chem., 283, 2008
|
|
4RHR
 
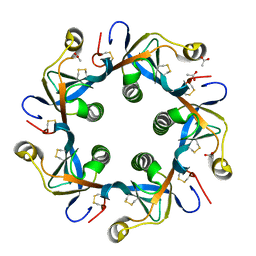 | | Crystal structure of PltB | | Descriptor: | ACETATE ION, Putative pertussis-like toxin subunit | | Authors: | Gao, X, Wang, J, Galan, J. | | Deposit date: | 2014-10-02 | | Release date: | 2014-10-29 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Host adaptation of a bacterial toxin from the human pathogen salmonella typhi.
Cell(Cambridge,Mass.), 159, 2014
|
|
4RHS
 
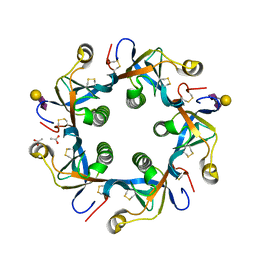 | | Crystal structure of GD2 bound PltB | | Descriptor: | ACETATE ION, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, Putative pertussis-like toxin subunit | | Authors: | Gao, X, Wang, J, Galan, J. | | Deposit date: | 2014-10-02 | | Release date: | 2014-10-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9211 Å) | | Cite: | Host adaptation of a bacterial toxin from the human pathogen salmonella typhi.
Cell(Cambridge,Mass.), 159, 2014
|
|
6DKJ
 
 | | human GIPR ECD and Fab complex | | Descriptor: | 1,2-ETHANEDIOL, Fab heavy chain, Fab light chain, ... | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Anti-obesity effects of GIPR antagonists alone and in combination with GLP-1R agonists in preclinical models.
Sci Transl Med, 10, 2018
|
|
5WHV
 
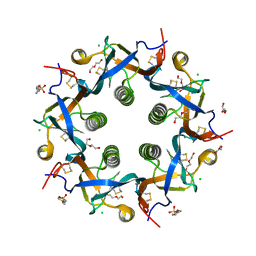 | | Crystal structure of ArtB | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ArtB protein, CALCIUM ION, ... | | Authors: | Gao, X, Galan, J.E. | | Deposit date: | 2017-07-18 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Evolution of host adaptation in the Salmonella typhoid toxin.
Nat Microbiol, 2, 2017
|
|
2F8P
 
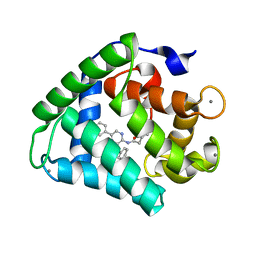 | | Crystal structure of obelin following Ca2+ triggered bioluminescence suggests neutral coelenteramide as the primary excited state | | Descriptor: | CALCIUM ION, N-[3-BENZYL-5-(4-HYDROXYPHENYL)PYRAZIN-2-YL]-2-(4-HYDROXYPHENYL)ACETAMIDE, Obelin | | Authors: | Liu, Z.J, Stepanyuk, G.A, Vysotski, E.S, Lee, J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-12-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of obelin after Ca2+-triggered bioluminescence suggests neutral coelenteramide as the primary excited state.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6NIR
 
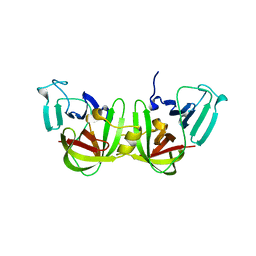 | | Crystal structure of a GII.4 norovirus HOV protease | | Descriptor: | HOV protease, HOV protease fragment | | Authors: | Prasad, B.V.V, Hu, L. | | Deposit date: | 2018-12-31 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | GII.4 Norovirus Protease Shows pH-Sensitive Proteolysis with a Unique Arg-His Pairing in the Catalytic Site.
J. Virol., 93, 2019
|
|
8SBJ
 
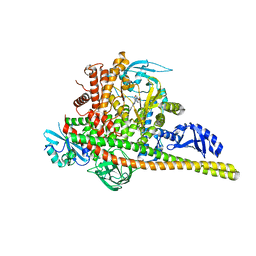 | | Co-structure Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform complexed with brain penetrant inhibitors | | Descriptor: | (2M)-7-[(3R)-3-methylmorpholin-4-yl]-5-[(3S)-3-methylmorpholin-4-yl]-2-(1H-pyrazol-3-yl)-3H-imidazo[4,5-b]pyridine, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Elling, R.A, Tang, J. | | Deposit date: | 2023-04-03 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Identification of Brain-Penetrant ATP-Competitive mTOR Inhibitors for CNS Syndromes.
J.Med.Chem., 66, 2023
|
|
8SBC
 
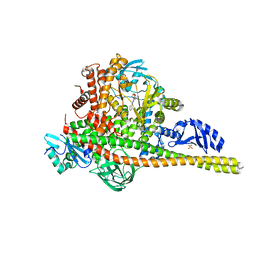 | | Co-structure of Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform and brain penetrant inhibitors | | Descriptor: | (2M)-7-[(3R)-3-methylmorpholin-4-yl]-5-[(3S)-3-methylmorpholin-4-yl]-2-(pyridin-2-yl)-1H-imidazo[4,5-b]pyridine, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Knapp, M.S, Elling, R.A, Tang, J. | | Deposit date: | 2023-04-03 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Brain-Penetrant ATP-Competitive mTOR Inhibitors for CNS Syndromes.
J.Med.Chem., 66, 2023
|
|
3QT3
 
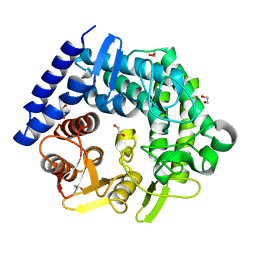 | | Analysis of a New Family of Widely Distributed Metal-independent alpha-Mannosidases Provides Unique Insight into the Processing of N-linked Glycans, Clostridium perfringens CPE0426 apo-structure | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein CPE0426 | | Authors: | Gregg, K.J, Zandberg, W.F, Hehemann, J.-H, Whitworth, G.E, Deng, L.E, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-22 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Analysis of a New Family of Widely Distributed Metal-independent {alpha}-Mannosidases Provides Unique Insight into the Processing of N-Linked Glycans.
J.Biol.Chem., 286, 2011
|
|
3QSP
 
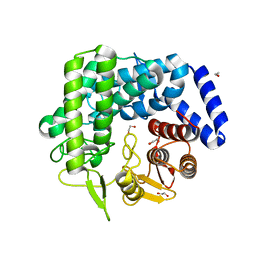 | | Analysis of a new family of widely distributed metal-independent alpha mannosidases provides unique insight into the processing of N-linked glycans, Streptococcus pneumoniae SP_2144 non-productive substrate complex with alpha-1,6-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose | | Authors: | Gregg, K.J, Zandberg, W.F, Hehemann, J.-H, Whitworth, G.E, Deng, L.E, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-21 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of a New Family of Widely Distributed Metal-independent {alpha}-Mannosidases Provides Unique Insight into the Processing of N-Linked Glycans.
J.Biol.Chem., 286, 2011
|
|
