6QFA
 
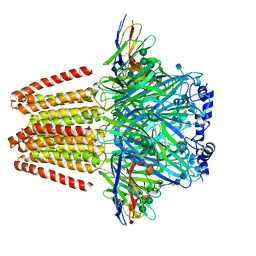 | | CryoEM structure of a beta3K279T GABA(A)R homomer in complex with histamine and megabody Mb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit beta-3,Gamma-aminobutyric acid receptor subunit beta-3, HISTAMINE, ... | | Authors: | Uchanski, T, Masiulis, S, Fischer, B, Kalichuk, V, Wohlkoening, A, Zoegg, T, Remaut, H, Vranken, W, Aricescu, A.R, Pardon, E, Steyaert, J. | | Deposit date: | 2019-01-09 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM.
Nat.Methods, 18, 2021
|
|
8PK8
 
 | |
8PKA
 
 | |
8PK9
 
 | |
7ZDM
 
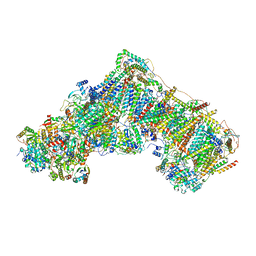 | | Complex I from Ovis aries at pH5.5, Closed state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Sazanov, L, Petrova, O. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7ZEB
 
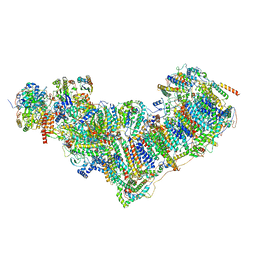 | | Complex I from Ovis aries at pH9, Closed state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Sazanov, L, Petrova, O. | | Deposit date: | 2022-03-30 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7ZDP
 
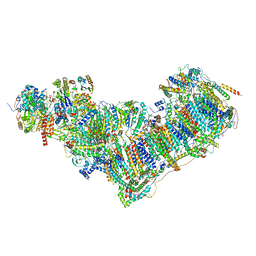 | | Complex I from Ovis aries at pH9, Open state | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE MONOPHOSPHATE, Acyl carrier protein, ... | | Authors: | Sazanov, L, Petrova, O. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7ZD6
 
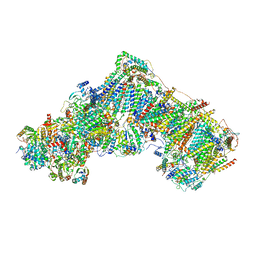 | | Complex I from Ovis aries, at pH7.4, Open state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Sazanov, L, Petrova, O. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7ZDJ
 
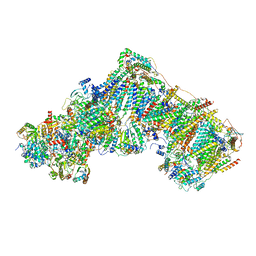 | | Complex I from Ovis aries at pH5.5, Open state | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Acyl carrier protein, Complex I subunit B13, ... | | Authors: | Petrova, O, Sazanov, L. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7ZDH
 
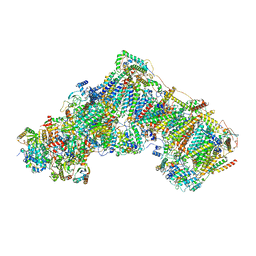 | | Complex I from Ovis aries at pH7.4, Closed state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Sazanov, L, Petrova, O. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
2W9R
 
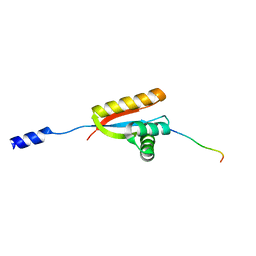 | | Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS | | Descriptor: | ATP-DEPENDENT CLP PROTEASE ADAPTER PROTEIN CLPS, DNA PROTECTION DURING STARVATION PROTEIN | | Authors: | Schuenemann, V, Kralik, S.M, Albrecht, R, Spall, S.K, Truscott, K.N, Dougan, D.A, Zeth, K. | | Deposit date: | 2009-01-28 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of N-End Rule Substrate Recognition in Escherichia Coli by the Clpap Adaptor Protein Clps.
Embo Rep., 10, 2009
|
|
6ORR
 
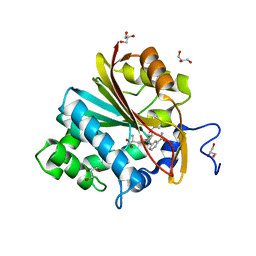 | | Co-crystal structure of human NicotinamideN-Methyltransferase (NNMT) in complex with High-Affinity Alkynyl Bisubstrate Inhibitor NS1 | | Descriptor: | 9-{9-amino-6-[(3-carbamoylphenyl)ethynyl]-5,6,7,8,9-pentadeoxy-D-glycero-alpha-L-talo-decofuranuronosyl}-9H-purin-6-amine, GLYCEROL, NNMT protein | | Authors: | May, E.J, Policarpo, R.L, Gaudet, R. | | Deposit date: | 2019-04-30 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High-Affinity Alkynyl Bisubstrate Inhibitors of NicotinamideN-Methyltransferase (NNMT).
J.Med.Chem., 62, 2019
|
|
1KSN
 
 | | Crystal Structure of Human Coagulation Factor XA Complexed with FXV673 | | Descriptor: | CALCIUM ION, COAGULATION FACTOR XA, METHYL-3-(4'-N-OXOPYRIDYLPHENOYL)-3-METHYL-2-(M-AMIDINOBENZYL)-PROPIONATE | | Authors: | Maignan, S, Guilloteau, J.P. | | Deposit date: | 2002-01-14 | | Release date: | 2002-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Optimization of the beta-aminoester class of factor Xa inhibitors. Part 2: Identification of FXV673 as a potent and selective inhibitor with excellent In vivo anticoagulant activity.
Bioorg.Med.Chem.Lett., 12, 2002
|
|
1SWK
 
 | | CORE-STREPTAVIDIN MUTANT W79F IN COMPLEX WITH BIOTIN AT PH 4.5 | | Descriptor: | BIOTIN, CORE-STREPTAVIDIN, EPI-BIOTIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWH
 
 | | CORE-STREPTAVIDIN MUTANT W79F AT PH 4.5 | | Descriptor: | CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWN
 
 | | CORE-STREPTAVIDIN MUTANT W108F IN COMPLEX WITH BIOTIN AT PH 7.0 | | Descriptor: | BIOTIN, CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWR
 
 | | CORE-STREPTAVIDIN MUTANT W120A IN COMPLEX WITH BIOTIN AT PH 7.5 | | Descriptor: | BIOTIN, CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWQ
 
 | | CORE-STREPTAVIDIN MUTANT W120A AT PH 7.5 | | Descriptor: | CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWP
 
 | | CORE-STREPTAVIDIN MUTANT W120F IN COMPLEX WITH BIOTIN AT PH 7.5 | | Descriptor: | BIOTIN, CORE-STREPTAVIDIN, EPI-BIOTIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWL
 
 | | CORE-STREPTAVIDIN MUTANT W108F AT PH 7.0 | | Descriptor: | CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWJ
 
 | | CORE-STREPTAVIDIN MUTANT W79F AT PH 4.5 | | Descriptor: | CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWO
 
 | | CORE-STREPTAVIDIN MUTANT W120F AT PH 7.5 | | Descriptor: | CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
6Z4C
 
 | | The structure of the N-terminal domain of RssB from E. coli | | Descriptor: | Regulator of RpoS | | Authors: | Zeth, K, Dimce, M, Terrence, D.M, Schuenemann, V, Dougan, D. | | Deposit date: | 2020-05-25 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into the RssB-Mediated Recognition and Delivery of sigma s to the AAA+ Protease, ClpXP.
Biomolecules, 10, 2020
|
|
4OV0
 
 | | Structure of Bacteriorhdopsin Transferred from Amphipol A8-35 to a Lipidic Mesophase | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Polovinkin, V, Gushchin, I, Sintsov, M, Round, E, Balandin, T, Chervakov, P, Schevchenko, V, Utrobin, P, Popov, A, Borshchevskiy, V, Mishin, A, Kuklin, A, Willbold, D, Popot, J.L, Gordeliy, V. | | Deposit date: | 2014-02-19 | | Release date: | 2014-10-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structure of a membrane protein transferred from amphipol to a lipidic mesophase.
J.Membr.Biol., 247, 2014
|
|
7BMH
 
 | | Crystal structure of a light-driven proton pump LR (Mac) from Leptosphaeria maculans | | Descriptor: | EICOSANE, OLEIC ACID, Opsin | | Authors: | Kovalev, K, Zabelskii, D, Dmitrieva, N, Volkov, O, Shevchenko, V, Astashkin, R, Zinovev, E, Gordeliy, V. | | Deposit date: | 2021-01-20 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based insights into evolution of rhodopsins.
Commun Biol, 4, 2021
|
|
