1BIK
 
 | | X-RAY STRUCTURE OF BIKUNIN FROM THE HUMAN INTER-ALPHA-INHIBITOR COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BIKUNIN, SULFATE ION | | Authors: | Xu, Y, Carr, P.D, Guss, J.M, Ollis, D.L. | | Deposit date: | 1997-11-26 | | Release date: | 1999-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of bikunin from the inter-alpha-inhibitor complex: a serine protease inhibitor with two Kunitz domains.
J.Mol.Biol., 276, 1998
|
|
1PIL
 
 | |
1YPN
 
 | | REDUCED FORM HYDROXYMETHYLBILANE SYNTHASE (K59Q MUTANT) CRYSTAL STRUCTURE AFTER 2 HOURS IN A FLOW CELL DETERMINED BY TIME-RESOLVED LAUE DIFFRACTION | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, HYDROXYMETHYLBILANE SYNTHASE | | Authors: | Helliwell, J.R, Nieh, Y.P, Raftery, J, Cassetta, A, Habash, J, Carr, P.D, Ursby, T, Wulff, M, Thompson, A.W, Niemann, A.C, Haedener, A. | | Deposit date: | 1998-06-26 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Time-Resolved Structures of Hydroxymethylbilane Synthase (Lys59Gln Mutant) as It Isloaded with Substrate in the Crystal Determined by Laue Diffraction
J.Chem.Soc.,Faraday Trans., 94, 1998
|
|
2DXL
 
 | |
2DXN
 
 | |
6CT3
 
 | | Directed evolutionary changes in Kemp Eliminase KE07 - Crystal 27 round 7-2 | | Descriptor: | 5-nitro-2-oxidanyl-benzenecarbonitrile, GLYCEROL, Kemp eliminase, ... | | Authors: | Jackson, C.J, Hong, N.-S, Carr, P.D. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The evolution of multiple active site configurations in a designed enzyme.
Nat Commun, 9, 2018
|
|
6C7T
 
 | |
6CAI
 
 | |
6C8B
 
 | |
6DKV
 
 | |
6DNJ
 
 | | Directed evolutionary changes in Kemp Eliminase KE07 - Crystal 28 round 5 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-nitro-1,2-benzoxazole, Kemp eliminase KE07 | | Authors: | Jackson, C.J, Hong, N.-S, Carr, P.D. | | Deposit date: | 2018-06-06 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The evolution of multiple active site configurations in a designed enzyme.
Nat Commun, 9, 2018
|
|
6DC1
 
 | | Directed evolutionary changes in Kemp Eliminase KE07 - Crystal 25 round 7 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-nitro-2-oxidanyl-benzenecarbonitrile, ... | | Authors: | Jackson, C.J, Hong, N.-S, Carr, P.D. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The evolution of multiple active site configurations in a designed enzyme.
Nat Commun, 9, 2018
|
|
6C7H
 
 | |
6C7M
 
 | |
6C7V
 
 | |
4FNG
 
 | | The alpha-esterase-7 carboxylesterase, E3, from the blowfly Lucilia cuprina | | Descriptor: | E3 alpha-esterase-7 caboxylesterase | | Authors: | Jackson, C.J, Liu, J.-W, Carr, P.D, Younis, F, Pandey, G, Coppin, C, Meirelles, T, Ollis, D.L, Tawfik, D.S, Weik, M, Oakeshott, J.G. | | Deposit date: | 2012-06-19 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and function of an insect alpha-carboxylesterase ( alpha Esterase7) associated with insecticide resistance.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4FNM
 
 | | The alpha-esterase-7 carboxylesterase, E3, from the blowfly Lucilia cuprina | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 alpha-esterase-7 carboxylesterase | | Authors: | Jackson, C.J, Liu, J.-W, Carr, P.D, Younis, F, Pandey, G, Coppin, C, Meirelles, T, Ollis, D.L, Tawfik, D.S, Weik, M, Oakeshott, J.G. | | Deposit date: | 2012-06-20 | | Release date: | 2013-12-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Structure and function of an insect alpha-carboxylesterase ( alpha Esterase7) associated with insecticide resistance.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3K7D
 
 | | C-terminal (adenylylation) domain of E.coli Glutamine Synthetase Adenylyltransferase | | Descriptor: | Glutamate-ammonia-ligase adenylyltransferase, SULFATE ION | | Authors: | Xu, Y, Carr, P.D, Vasudevan, S.G, Ollis, D.L. | | Deposit date: | 2009-10-12 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Adenylylation Domain of E. coli Glutamine Synthetase Adenylyl Transferase: Evidence for Gene Duplication and Evolution of a New Active Site.
J.Mol.Biol., 396, 2010
|
|
1QY7
 
 | | The structure of the PII protein from the cyanobacteria Synechococcus sp. PCC 7942 | | Descriptor: | NICKEL (II) ION, Nitrogen regulatory protein P-II, SULFATE ION | | Authors: | Xu, Y, Carr, P.D, Clancy, P, Garcia-Dominguez, M, Forchhammer, K, Florencio, F, Tandeau de Marsac, N, Vasudevan, S.G, Ollis, D.L. | | Deposit date: | 2003-09-09 | | Release date: | 2003-09-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of the PII proteins from the cyanobacteria Synechococcus sp. PCC 7942 and Synechocystis sp. PCC 6803.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4Z08
 
 | |
2D2H
 
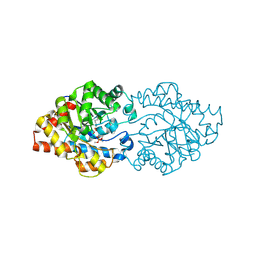 | | OpdA from Agrobacterium radiobacter with bound inhibitor trimethyl phosphate at 1.8 A resolution | | Descriptor: | COBALT (II) ION, TRIMETHYL PHOSPHATE, phosphotriesterase | | Authors: | Jackson, C, Kim, H.K, Carr, P.D, Liu, J.W, Ollis, D.L. | | Deposit date: | 2005-09-09 | | Release date: | 2005-09-20 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of an enzyme-product complex reveals the critical role of a terminal hydroxide nucleophile in the bacterial phosphotriesterase mechanism
Biochim.Biophys.Acta, 1752, 2005
|
|
1ZJ5
 
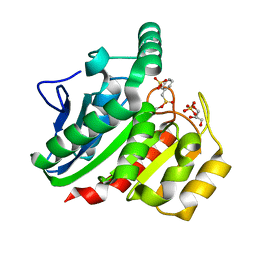 | | Crystal Structure Analysis of the dienelactone hydrolase mutant (E36D, C123S, A134S, S208G, A229V, K234R) bound with the PMS moiety of the protease inhibitor, Phenylmethylsulfonyl fluoride (PMSF)- 1.7 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-28 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4ZKY
 
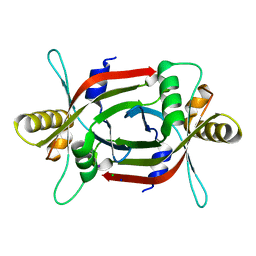 | | Structure of F420 binding protein, MSMEG_6526, from Mycobacterium smegmatis | | Descriptor: | CHLORIDE ION, IODIDE ION, Pyridoxamine 5-phosphate oxidase, ... | | Authors: | Lee, B.M, Carr, P.D, Ahmed, F.H, Jackson, C.J. | | Deposit date: | 2015-05-01 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Sequence-Structure-Function Classification of a Catalytically Diverse Oxidoreductase Superfamily in Mycobacteria.
J.Mol.Biol., 427, 2015
|
|
5BNC
 
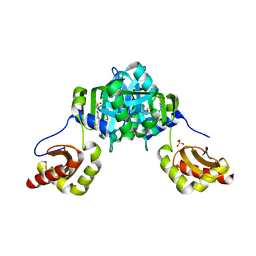 | | Structure of heme binding protein MSMEG_6519 from Mycobacterium smegmatis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Ahmed, F.H, Carr, P.D, Jackson, C.J. | | Deposit date: | 2015-05-26 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sequence-Structure-Function Classification of a Catalytically Diverse Oxidoreductase Superfamily in Mycobacteria.
J.Mol.Biol., 427, 2015
|
|
1ZIC
 
 | | Crystal Structure Analysis of the dienelactone hydrolase (C123S, R206A) mutant- 1.7 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
