8PT5
 
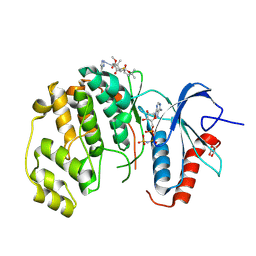 | | ERK2 covelently bound to RU187 cyclohexenone based inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Sok, P, Poti, A, Remenyi, A. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a precision-guided warhead scaffold.
Nat Commun, 15, 2024
|
|
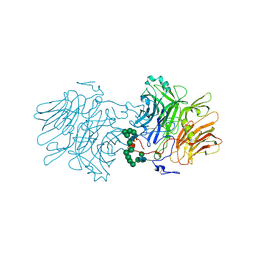
5FKB
 
 | |
9G15
 
 | | Crystal structure of marine actinobacteria clade rhodopsin (MAR) in the O* state, pH 8.8 | | Descriptor: | Bacteriorhodopsin, EICOSANE, OLEIC ACID, ... | | Authors: | Bukhdruker, S, Kovalev, K, Astashkin, R, Gordeliy, V. | | Deposit date: | 2024-07-09 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Proteorhodopsin insights into the molecular mechanism of vectorial proton transport.
Sci Adv, 11, 2025
|
|
9G16
 
 | | Crystal structure of marine actinobacteria clade rhodopsin (MAR) in the O state obtained by cryotrapping | | Descriptor: | Bacteriorhodopsin, EICOSANE, OLEIC ACID, ... | | Authors: | Bukhdruker, S, Kovalev, K, Astashkin, R, Gordeliy, V. | | Deposit date: | 2024-07-09 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Proteorhodopsin insights into the molecular mechanism of vectorial proton transport.
Sci Adv, 11, 2025
|
|
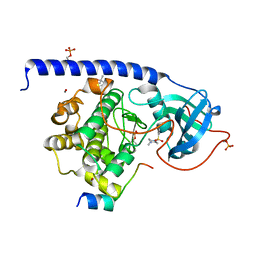
5LCT
 
 | |
8QWX
 
 | | Ligninolytic manganese peroxidase Ape-MnP1 from Agaricales mushrooms | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Santillana, E, Romero, A. | | Deposit date: | 2023-10-20 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-function characterization of two enzymes from novel subfamilies of manganese peroxidases secreted by the lignocellulose-degrading Agaricales fungi Agrocybe pediades and Cyathus striatus.
Biotechnol Biofuels Bioprod, 17, 2024
|
|
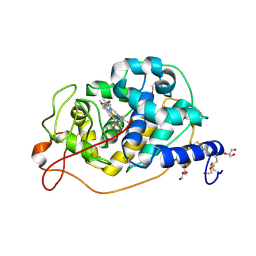
8QX0
 
 | |
8QWT
 
 | |
5LCU
 
 | |
5M71
 
 | |
5UR5
 
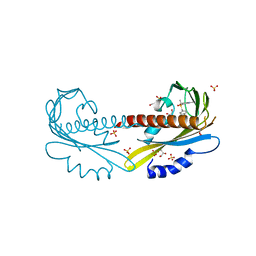 | | PYR1 bound to the rationally designed agonist 4m | | Descriptor: | Abscisic acid receptor PYR1, GLYCEROL, N-(4-cyano-3-ethyl-5-methylphenyl)-1-(4-methylphenyl)methanesulfonamide, ... | | Authors: | Peterson, F.C, Vaidya, A, Jensen, D.R, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2017-02-09 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A Rationally Designed Agonist Defines Subfamily IIIA Abscisic Acid Receptors As Critical Targets for Manipulating Transpiration.
ACS Chem. Biol., 12, 2017
|
|
5FMC
 
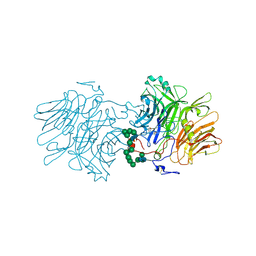 | | Structure of D80A-fructofuranosidase from Xanthophyllomyces dendrorhous complexed with fructose and BIS-TRIS propane buffer | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ramirez-Escudero, M, Sanz-Aparicio, J. | | Deposit date: | 2015-11-02 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural Analysis of Beta-Fructofuranosidase from Xanthophyllomyces Dendrorhous Reveals Unique Features and the Crucial Role of N-Glycosylation in Oligomerization and Activity
J.Biol.Chem., 291, 2016
|
|
5M6V
 
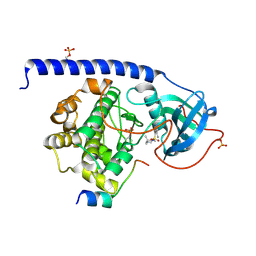 | |
5LCQ
 
 | |
8R1M
 
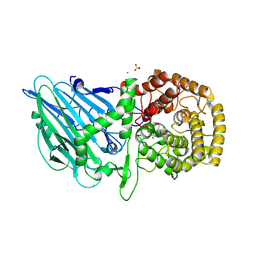 | | Structure of TxGH116 with covalently bound N-azido-octyl aziridine | | Descriptor: | (1R,2R,3R,4S)-4-[8-(azanyldiazenyl)octylamino]-3-(hydroxymethyl)cyclopentane-1,2-diol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Offen, W.A, Davies, G.J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective labelling of GBA2 in cells with fluorescent beta-d-arabinofuranosyl cyclitol aziridines.
Chem Sci, 15, 2024
|
|
5FKC
 
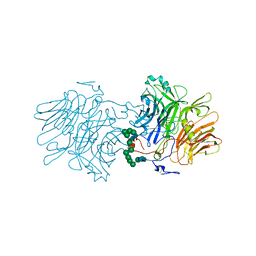 | |
5M0B
 
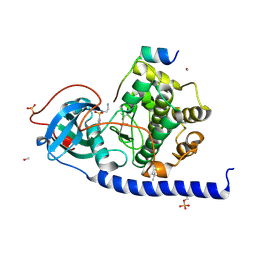 | |
5WRN
 
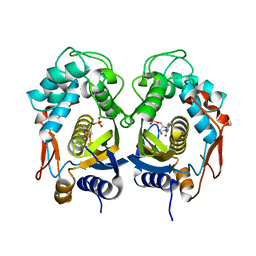 | |
9F41
 
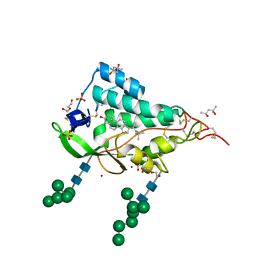 | | Crystal structure of the NTD domain from S. cerevisia Niemann-Pick type C protein NCR1 with cholesterol bound | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHOLESTEROL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nel, L, Olesen, E, Frain, K.M, Pedersen, B.P. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural and biochemical analysis of ligand binding in yeast Niemann-Pick type C1-related protein.
Life Sci Alliance, 8, 2025
|
|
9F40
 
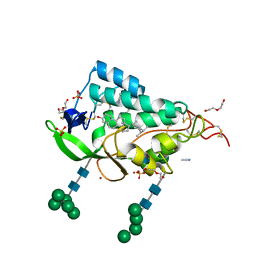 | | Crystal structure of the NTD domain from S. cerevisia Niemann-Pick type C protein NCR1 with ergosterol bound | | Descriptor: | ACETONITRILE, DI(HYDROXYETHYL)ETHER, ERGOSTEROL, ... | | Authors: | Nel, L, Olesen, E, Frain, K.M, Pedersen, B.P. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural and biochemical analysis of ligand binding in yeast Niemann-Pick type C1-related protein.
Life Sci Alliance, 8, 2025
|
|
1K1C
 
 | | Solution Structure of Crh, the Bacillus subtilis Catabolite Repression HPr | | Descriptor: | catabolite repression HPr-like protein | | Authors: | Favier, A, Brutscher, B, Blackledge, M, Galinier, A, Deutscher, J, Penin, F, Marion, D. | | Deposit date: | 2001-09-25 | | Release date: | 2001-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of Crh, the Bacillus subtilis catabolite repression HPr.
J.Mol.Biol., 317, 2002
|
|
9DI6
 
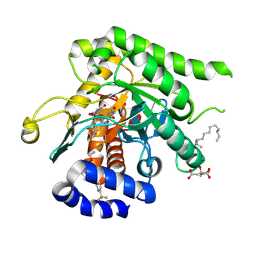 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM679 (ethyl 1,4-dimethyl-5-((6-(trifluoromethyl)pyridin-3-yl)methyl)-1H-pyrazole-3-carboxylate) | | Descriptor: | 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, CITRIC ACID, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Deng, X, Tomchick, D, Phillips, M. | | Deposit date: | 2024-09-05 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9DIK
 
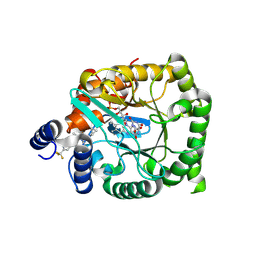 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM681 (N-cyclopropyl-1,4-dimethyl-5-((6-(trifluoromethyl)pyridin-3-yl)methyl)-1H-pyrazole-3-carboxamide) | | Descriptor: | 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Tomchick, D, Phillips, M. | | Deposit date: | 2024-09-05 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9DIZ
 
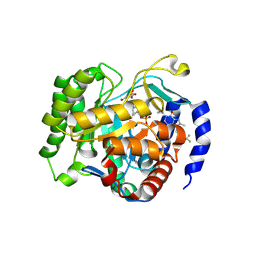 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM959 (6-cyclopropyl-2,4-dimethyl-3-((6-(trifluoromethyl)pyridin-3-yl)methyl)-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one) | | Descriptor: | 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, 6-cyclopropyl-2,4-dimethyl-3-{[6-(trifluoromethyl)pyridin-3-yl]methyl}-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Deng, X, Tomchick, D, Phillips, M. | | Deposit date: | 2024-09-06 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9DKY
 
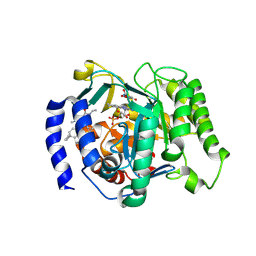 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM1174 (3-((7-azaspiro[3.5]nonan-7-yl)methyl)-4-cyclopropyl-6-ethyl-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one) | | Descriptor: | 3-[(7-azaspiro[3.5]nonan-7-yl)methyl]-4-cyclopropyl-6-ethyl-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one, 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Deng, X, Tomchick, D, Phillips, M. | | Deposit date: | 2024-09-10 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
