4Y40
 
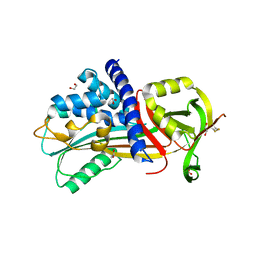 | | Structure of Vaspin mutant D305C V383C | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Serpin A12 | | Authors: | Pippel, J, Strater, N, Ulbricht, D, Schultz, S, Meier, R, Heiker, J.T. | | Deposit date: | 2015-02-10 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A unique serpin P1' glutamate and a conserved beta-sheet C arginine are key residues for activity, protease recognition and stability of serpinA12 (vaspin).
Biochem.J., 470, 2015
|
|
8DLI
 
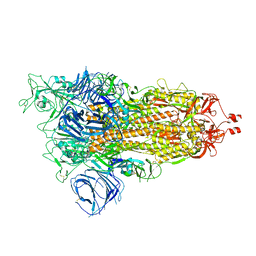 | | Cryo-EM structure of SARS-CoV-2 Alpha (B.1.1.7) spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
4Y4O
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome with rRNA modifications and bound to protein Y (YfiA) at 2.3A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Polikanov, Y.S, Melnikov, S.V, Soll, D, Steitz, T.A. | | Deposit date: | 2015-02-10 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8DLQ
 
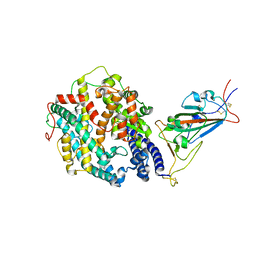 | | Cryo-EM structure of SARS-CoV-2 Gamma (P.1) spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
1GQJ
 
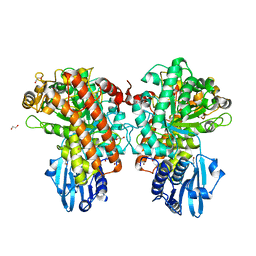 | | Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with xylobiose | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-D-GLUCURONIDASE, COBALT (II) ION, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structural Basis for Catalysis and Specificity of the Pseudomonas Cellulosa Alpha-Glucuronidase, Glca67A
Structure, 10, 2002
|
|
8DLZ
 
 | | Cryo-EM structure of SARS-CoV-2 D614G spike protein in complex with VH ab6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
4YE0
 
 | | Stress-induced protein 1 truncation mutant (43 - 140) from Caenorhabditis elegans | | Descriptor: | SULFATE ION, Stress-induced protein 1 | | Authors: | Fleckenstein, T, Kastenmueller, A, Stein, M.L, Peters, C, Daake, M, Krause, M, Weinfurtner, D, Haslbeck, M, Weinkauf, S, Groll, M, Buchner, J. | | Deposit date: | 2015-02-23 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Chaperone Activity of the Developmental Small Heat Shock Protein Sip1 Is Regulated by pH-Dependent Conformational Changes.
Mol.Cell, 58, 2015
|
|
6Q7D
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with the NVP-BHG712 derivative ATDL13 | | Descriptor: | 3-[[4-imidazol-1-yl-6-(4-oxidanylpiperidin-1-yl)-1,3,5-triazin-2-yl]amino]-4-methyl-~{N}-[3-(trifluoromethyl)phenyl]benzamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Gande, S.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-12-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.978 Å) | | Cite: | Effects of NVP-BHG712 chemical modifications on EPHA2 binding and affinity
To Be Published
|
|
1RQC
 
 | | Crystals of peptide deformylase from Plasmodium falciparum with ten subunits per asymmetric unit reveal critical characteristics of the active site for drug design | | Descriptor: | COBALT (II) ION, formylmethionine deformylase | | Authors: | Robien, M.A, Nguyen, K.T, Kumar, A, Hirsh, I, Turley, S, Pei, D, Hol, W.G. | | Deposit date: | 2003-12-04 | | Release date: | 2004-01-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An improved crystal form of Plasmodium falciparum peptide deformylase
Protein Sci., 13, 2004
|
|
6QA0
 
 | | MSRB3 - AA 1-137 | | Descriptor: | CHLORIDE ION, Methionine-R-sulfoxide reductase B3, SULFATE ION, ... | | Authors: | Javitt, G, Fass, D. | | Deposit date: | 2018-12-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Structure and Electron-Transfer Pathway of the Human Methionine Sulfoxide Reductase MsrB3.
Antioxid.Redox Signal., 33, 2020
|
|
6QAB
 
 | | Human Butyrylcholinesterase in complex with (S)-N-(1-((2-cycloheptylethyl)amino)-3-(1H-indol-3-yl)-1-oxopropan-2-yl)-N,N-dimethylbutan-1-aminium | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Nachon, F, Harst, M, Knez, D, Gobec, S. | | Deposit date: | 2018-12-19 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Tryptophan-derived butyrylcholinesterase inhibitors as promising leads against Alzheimer's disease.
Chem.Commun.(Camb.), 55, 2019
|
|
6QAL
 
 | | ERK2 mini-fragment binding | | Descriptor: | 1,1-bis(oxidanylidene)thietan-3-ol, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-19 | | Release date: | 2019-03-27 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
1GPF
 
 | |
6QAQ
 
 | | ERK2 mini-fragment binding | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, thiophen-3-ylmethylazanium | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-19 | | Release date: | 2019-03-27 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
6QAW
 
 | | ERK2 mini-fragment binding | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, [1-(7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-yl]methylazanium | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-19 | | Release date: | 2019-03-27 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
8GV9
 
 | | The cryo-EM structure of hAE2 with chloride ion | | Descriptor: | Anion exchange protein 2, CHLORIDE ION | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
4L3B
 
 | | X-ray structure of the HRV2 A particle uncoating intermediate | | Descriptor: | Protein VP1, Protein VP2, Protein VP3 | | Authors: | Vives-Adrian, L, Querol-Audi, J, Garriga, D, Pous, J, Verdaguer, N. | | Deposit date: | 2013-06-05 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Uncoating of common cold virus is preceded by RNA switching as determined by X-ray and cryo-EM analyses of the subviral A-particle.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8GVA
 
 | | The intermediate structure of hAE2 in basic pH | | Descriptor: | Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVE
 
 | | The asymmetry structure of hAE2 | | Descriptor: | Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
1RQP
 
 | | Crystal structure and mechanism of a bacterial fluorinating enzyme | | Descriptor: | 5'-fluoro-5'-deoxyadenosine synthase, S-ADENOSYLMETHIONINE | | Authors: | Dong, C, Huang, F, Deng, H, Schaffrath, C, Spencer, J.B, O'Hagan, D, Naismith, J.H. | | Deposit date: | 2003-12-06 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and mechanism of a bacterial fluorinating enzyme
Nature, 427, 2004
|
|
8GV8
 
 | | The cryo-EM structure of hAE2 with DIDS | | Descriptor: | 2,2'-ethane-1,2-diylbis{5-[(sulfanylmethyl)amino]benzenesulfonic acid}, Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GOK
 
 | |
4YYS
 
 | | Ficin B crystal form II | | Descriptor: | Ficin isoform B, PHOSPHATE ION | | Authors: | Azarkan, M, Baeyens-Volant, D, Loris, R. | | Deposit date: | 2015-03-24 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of four variants of the protease Ficin from the common fig
To Be Published
|
|
8GAE
 
 | | Hsp90 provides platform for CRaf dephosphorylation by PP5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, ... | | Authors: | Jaime-Garza, M, Nowotny, C.A, Coutandin, D, Wang, F, Tabios, M, Agard, D.A. | | Deposit date: | 2023-02-22 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Hsp90 provides a platform for kinase dephosphorylation by PP5.
Nat Commun, 14, 2023
|
|
8GXD
 
 | | L-LEUCINE DEHYDROGENASE FROM EXIGUOBACTERIUM SIBIRICUM | | Descriptor: | CALCIUM ION, GLYCEROL, Glu/Leu/Phe/Val dehydrogenase | | Authors: | Mu, X, Nie, Y, Wu, T, Wang, Y, Zhang, N, Yin, D, Xu, Y. | | Deposit date: | 2022-09-19 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Reshaping Substrate-Binding Pocket of Leucine Dehydrogenase for Bidirectionally Accessing Structurally Diverse Substrates
Acs Catalysis, 13, 2023
|
|
