2Z85
 
 | | Ligand Migration and Binding in The Dimeric Hemoglobin of Scapharca Inaequivalvis: M37F Unliganded | | Descriptor: | Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Royer Jr, W.E, Nienhaus, K, Palladino, P, Nienhaus, G.U. | | Deposit date: | 2007-09-02 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ligand Migration and Binding in the Dimeric Hemoglobin of Scapharca inaequivalvis
Biochemistry, 46, 2007
|
|
3BL6
 
 | | Crystal structure of Staphylococcus aureus 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase in complex with formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, 5'-methylthioadenosine nucleosidase/S-adenosylhomocysteine nucleosidase | | Authors: | Siu, K.K.W, Lee, J.E, Smith, G.D, Horvatin, C, Howell, P.L. | | Deposit date: | 2007-12-10 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Staphylococcus aureus 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase
Acta Crystallogr.,Sect.F, 64, 2008
|
|
3B91
 
 | | Minimally Hinged Hairpin Ribozyme Incorporates Ade38(2AP) and 2',5'-Phosphodiester Linkage Mutations at the Active Site | | Descriptor: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-11-02 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3B58
 
 | | Minimally Junctioned Hairpin Ribozyme Incorporates A38G Mutation and a 2',5'-Phosphodiester Linkage at the Active Site | | Descriptor: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-10-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3B39
 
 | |
3B5S
 
 | | Minimally Hinged Hairpin Ribozyme Incorporates A38DAP Mutation and 2'-O-methyl Modification at the Active Site | | Descriptor: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-10-26 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3C79
 
 | | Crystal structure of Aplysia californica AChBP in complex with the neonicotinoid imidacloprid | | Descriptor: | (2E)-1-[(6-chloropyridin-3-yl)methyl]-N-nitroimidazolidin-2-imine, ISOPROPYL ALCOHOL, Soluble acetylcholine receptor | | Authors: | Talley, T.T, Harel, M, Hibbs, R.E, Tomizawa, M, Casida, J.E, Taylor, P.W. | | Deposit date: | 2008-02-06 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Atomic interactions of neonicotinoid agonists with AChBP: molecular recognition of the distinctive electronegative pharmacophore.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3C84
 
 | | Crystal structure of a complex of AChBP from aplysia californica and the neonicotinoid thiacloprid | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, Soluble acetylcholine receptor, ... | | Authors: | Talley, T.T, Harel, M, Hibbs, R.E, Tomizawa, M, Casida, J.E, Taylor, P.W. | | Deposit date: | 2008-02-08 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Atomic interactions of neonicotinoid agonists with AChBP: molecular recognition of the distinctive electronegative pharmacophore.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3C8E
 
 | | Crystal Structure Analysis of yghU from E. Coli | | Descriptor: | GLUTATHIONE, yghU, glutathione S-transferase homologue | | Authors: | Harp, J, Ladner, J.E, Schaab, M.R, Stourman, N.V, Armstrong, R.N. | | Deposit date: | 2008-02-11 | | Release date: | 2009-02-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Function of YghU, a Nu-Class Glutathione Transferase Related to YfcG from Escherichia coli.
Biochemistry, 50, 2011
|
|
3B1T
 
 | | Crystal structure of human peptidylarginine deiminase 4 in complex with o-Cl-amidine | | Descriptor: | 2-{[(2S)-1-amino-5-{[(1Z)-2-chloroethanimidoyl]amino}-1-oxopentan-2-yl]carbamoyl}benzoic acid, CALCIUM ION, Protein-arginine deiminase type-4, ... | | Authors: | Causey, C.P, Jones, J.E, Slack, J.L, Kamei, D, Jones Jr, L.E, Subramanian, V, Knuckley, B, Ebrahimi, P, Chumanevich, A.A, Luo, Y, Hashimoto, H, Shimizu, T, Sato, M, Hofseth, L.J, Thompson, P.R. | | Deposit date: | 2011-07-13 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Development of N-alpha-(2-Carboxyl)benzoyl-N(5)-(2-fluoro-1-iminoethyl)-l-ornithine Amide (o-F-amidine) and N-alpha-(2-Carboxyl)benzoyl-N(5)-(2-chloro-1-iminoethyl)-l-ornithine Amide (o-Cl-amidine) As Second Generation Protein Arginine Deiminase (PAD) Inhibitors
J.Med.Chem., 54, 2011
|
|
2YCF
 
 | | Crystal Structure of Checkpoint Kinase 2 in complex with Inhibitor PV1531 | | Descriptor: | (2E)-N-hydroxy-2-[1-(4-{[(4-{(1E)-1-[2-(N'-hydroxycarbamimidoyl)hydrazinylidene]ethyl}phenyl)carbamoyl]amino}phenyl)ethylidene]hydrazinecarboximidamide, MAGNESIUM ION, NITRATE ION, ... | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C.R, Pommier, Y, Shoemaker, R.H, Zhang, G, Waugh, D.S. | | Deposit date: | 2011-03-14 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Characterization of Inhibitor Complexes with Checkpoint Kinase 2 (Chk2), a Drug Target for Cancer Therapy.
J.Struct.Biol., 176, 2011
|
|
3C8P
 
 | | X-ray structure of Viscotoxin A1 from Viscum album L. | | Descriptor: | Viscotoxin A1 | | Authors: | Pal, A, Debreczeni, J.E, Sevvana, M, Gruene, T, Kahle, B, Zeeck, A, Sheldrick, G.M. | | Deposit date: | 2008-02-13 | | Release date: | 2008-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structures of viscotoxins A1 and B2 from European mistletoe solved using native data alone
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3BZN
 
 | | Crystal Structure of Open form of Menaquinone-Specific Isochorismate Synthase, MenF | | Descriptor: | MAGNESIUM ION, Menaquinone-specific isochorismate synthase, SULFATE ION | | Authors: | Parsons, J.F, Shi, K.M, Ladner, J.E. | | Deposit date: | 2008-01-18 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of isochorismate synthase in complex with magnesium.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3CG5
 
 | |
3CSY
 
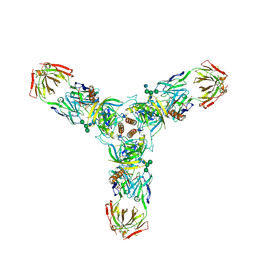 | | Crystal structure of the trimeric prefusion Ebola virus glycoprotein in complex with a neutralizing antibody from a human survivor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein GP1, ... | | Authors: | Lee, J.E, Fusco, M.L, Hessell, A.J, Oswald, W.B, Burton, D.R, Saphire, E.O. | | Deposit date: | 2008-04-10 | | Release date: | 2008-07-08 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor.
Nature, 454, 2008
|
|
3CR1
 
 | | crystal structure of a minimal, mutant, all-RNA hairpin ribozyme (A38C, A-1OMA) grown from MgCl2 | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*G)-3'), RNA (5'-R(*UP*CP*GP*UP*GP*GP*UP*CP*CP*AP*UP*UP*AP*CP*CP*UP*GP*CP*C)-3'), ... | | Authors: | Salter, J.D, Wedekind, J.E. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3B5A
 
 | | Crystal Structure of a Minimally Hinged Hairpin Ribozyme Incorporating A38G mutation with a 2'OMe modification at the active site | | Descriptor: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-10-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3D7J
 
 | | SCO6650, a 6-pyruvoyltetrahydropterin synthase homolog from Streptomyces coelicolor | | Descriptor: | CHLORIDE ION, SODIUM ION, Uncharacterized protein SCO6650 | | Authors: | Spoonamore, J.E, Roberts, S.A, Heroux, A, Bandarian, V. | | Deposit date: | 2008-05-21 | | Release date: | 2008-10-21 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a 6-pyruvoyltetrahydropterin synthase homolog from Streptomyces coelicolor.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
3CP6
 
 | | Crystal structure of human farnesyl diphosphate synthase (T201A mutant) complexed with Mg and biphosphonate inhibitor | | Descriptor: | (4aS,7aR)-octahydro-1H-cyclopenta[b]pyridine-6,6-diylbis(phosphonic acid), Farnesyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Pilka, E.S, Dunford, J.E, Guo, K, Pike, A.C.W, von Delft, F, Barnett, B.L, Ebetino, F.H, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Russell, R.G.G, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Human farnesyl diphosphate synthase (T201A mutant) complexed with Mg and biphosphonate inhibitor.
To be Published
|
|
1SHC
 
 | | SHC PTB DOMAIN COMPLEXED WITH A TRKA RECEPTOR PHOSPHOPEPTIDE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | SHC, TRKA RECEPTOR PHOSPHOPEPTIDE | | Authors: | Zhou, M.-M, Ravichandran, K.S, Olejniczak, E.T, Petros, A.M, Meadows, R.P, Sattler, M, Harlan, J.E, Wade, W.S, Burakoff, S.J, Fesik, S.W. | | Deposit date: | 1996-03-27 | | Release date: | 1997-05-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure and ligand recognition of the phosphotyrosine binding domain of Shc.
Nature, 378, 1995
|
|
1TJ5
 
 | | X-Ray structure of the Sucrose-Phosphatase (SPP) from Synechocystis sp. PCC6803 in complex with sucrose and phosphate | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Sucrose-Phosphatase, ... | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.-L. | | Deposit date: | 2004-06-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell.
Plant Cell, 17, 2005
|
|
1U7T
 
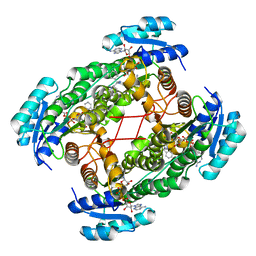 | | Crystal Structure of ABAD/HSD10 with a Bound Inhibitor | | Descriptor: | 1-AZEPAN-1-YL-2-PHENYL-2-(4-THIOXO-1,4-DIHYDRO-PYRAZOLO[3,4-D]PYRIMIDIN-5-YL)ETHANONE ADDUCT, 3-hydroxyacyl-CoA dehydrogenase type II, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kissinger, C.R, Rejto, P.A, Pelletier, L.A, Showalter, R.E, Villafranca, J.E. | | Deposit date: | 2004-08-04 | | Release date: | 2004-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human ABAD/HSD10 with a bound inhibitor: implications for design of Alzheimer's disease therapeutics
J.Mol.Biol., 342, 2004
|
|
1U2T
 
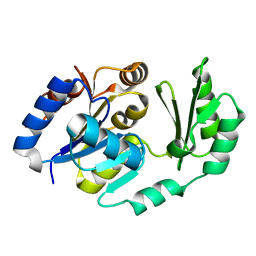 | | X-Ray structure of the sucrose-phosphatase (SPP) from Synechocystis sp. PCC6803 in complex with sucrose6P | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, sucrose-phosphatase (SPP) | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.-L. | | Deposit date: | 2004-07-20 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell
PLANT CELL, 17, 2005
|
|
1TY9
 
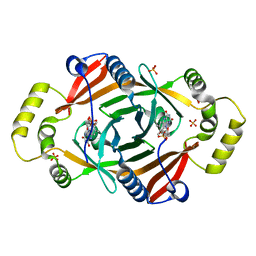 | | X-RAY CRYSTAL STRUCTURE OF PHZG FROM PSEUDOMONAS FLUORESCENS | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phenazine biosynthesis protein phzG, SULFATE ION | | Authors: | Parsons, J.F, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2004-07-07 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the phenazine biosynthesis enzyme PhzG.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5W90
 
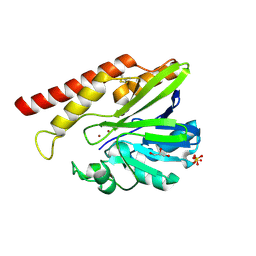 | | FEZ-1 metallo-beta-lactamase from Legionella gormanii modelled with unknown ligand | | Descriptor: | FEZ-1 protein, GLYCEROL, SULFATE ION, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Kahn, R, Shabalin, I.G, Raczynska, J.E, Jaskolski, M, Minor, W, Papamicael, C, Frere, J.M, Galleni, M, Dideberg, O. | | Deposit date: | 2017-06-22 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Three-dimensional structure of FEZ-1, a monomeric subclass B3 metallo-beta-lactamase from Fluoribacter gormanii, in native form and in complex with D-captopril.
J. Mol. Biol., 325, 2003
|
|
