5ZUQ
 
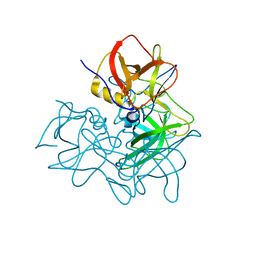 | | P domain of GII.17-1978 | | Descriptor: | VP1 | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-08 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5WJ5
 
 | |
8YAK
 
 | | Cryo-EM structure and rational engineering of a novel efficient ochratoxin A-detoxifying amidohydrolase | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Imidazolonepropionase, ZINC ION | | Authors: | Dai, L.H, Xu, Y.H, Hu, Y.M, Niu, D, Yang, X.C, Shen, P.P, Li, X, Xie, Z.Z, Li, H, Guo, R.-T, Chen, C.-C. | | Deposit date: | 2024-02-09 | | Release date: | 2024-12-18 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Functional characterization and structural basis of an efficient ochratoxin A-degrading amidohydrolase.
Int.J.Biol.Macromol., 278, 2024
|
|
7URF
 
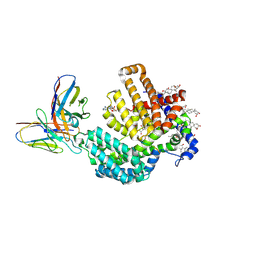 | | Human HHAT H379C in complex with SHH N-terminal peptide | | Descriptor: | 3H02 heavy chain, 3H02 light chain, Digitonin, ... | | Authors: | Liu, Y, Qi, X, Li, X. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-20 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanisms and inhibition of Porcupine-mediated Wnt acylation.
Nature, 607, 2022
|
|
7URE
 
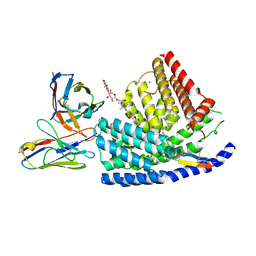 | | Human PORCN in complex with palmitoleoylated WNT3A peptide | | Descriptor: | 2C11 heavy chain, 2C11 light chain, Digitonin, ... | | Authors: | Liu, Y, Qi, X, Li, X. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Mechanisms and inhibition of Porcupine-mediated Wnt acylation.
Nature, 607, 2022
|
|
7URA
 
 | | Human PORCN in complex with Palmitoleoyl-CoA | | Descriptor: | 2C11 heavy chain, 2C11 light chain, CHOLESTEROL, ... | | Authors: | Liu, Y, Qi, X, Li, X. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Mechanisms and inhibition of Porcupine-mediated Wnt acylation.
Nature, 607, 2022
|
|
5ZUS
 
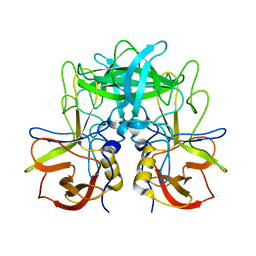 | | P domain of GII.17-2014/15 | | Descriptor: | VP1 | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-08 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5XRA
 
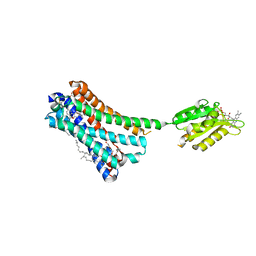 | | Crystal structure of the human CB1 in complex with agonist AM11542 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6aR,10aR)-3-(8-bromanyl-2-methyl-octan-2-yl)-6,6,9-trimethyl-6a,7,10,10a-tetrahydrobenzo[c]chromen-1-ol, CHOLESTEROL, ... | | Authors: | Hua, T, Vemuri, K, Nikas, P.S, Laprairie, R.B, Wu, Y, Qu, L, Pu, M, Korde, A, Shan, J, Ho, J.H, Han, G.W, Ding, K, Li, X, Liu, H, Hanson, M.A, Zhao, S, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of agonist-bound human cannabinoid receptor CB1
Nature, 547, 2017
|
|
4J1G
 
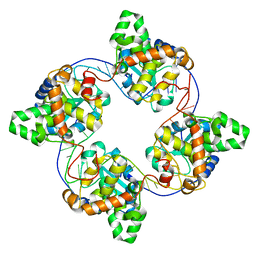 | | Leanyer orthobunyavirus nucleoprotein-ssRNA complex | | Descriptor: | Nucleocapsid, RNA (45-MER) | | Authors: | Niu, F, Shaw, N, Wang, Y, Jiao, L, Ding, W, Li, X, Zhu, P, Upur, H, Ouyang, S, Cheng, G, Liu, Z.J. | | Deposit date: | 2013-02-01 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Structure of the Leanyer orthobunyavirus nucleoprotein-RNA complex reveals unique architecture for RNA encapsidation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KUC
 
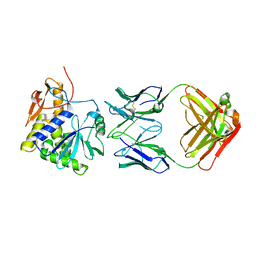 | |
7XL5
 
 | | Crystal structure of the H42T/A85G/I86A mutant of a nadp-dependent alcohol dehydrogenase | | Descriptor: | NADP-dependent isopropanol dehydrogenase | | Authors: | Jiang, Y.Y, Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2022-04-21 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Engineering the hydrogen transfer pathway of an alcohol dehydrogenase to increase activity by rational enzyme design
Mol Catal, 530, 2022
|
|
7C4U
 
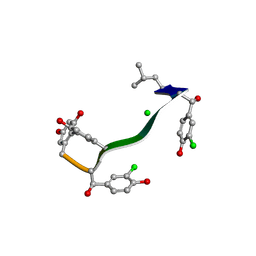 | | MicroED structure of orthorhombic Vancomycin at 1.2 A resolution | | Descriptor: | CHLORIDE ION, Vancomycin, vancosamine-(1-2)-beta-D-glucopyranose | | Authors: | Fan, Q, Zhou, H, Li, X, Wang, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-08-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.2 Å) | | Cite: | Precise Control Over Kinetics of Molecular Assembly: Production of Particles with Tunable Sizes and Crystalline Forms.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7C4V
 
 | | MicroED structure of anorthic Vancomycin at 1.05 A resolution | | Descriptor: | CHLORIDE ION, Vancomycin, vancosamine-(1-2)-beta-D-glucopyranose | | Authors: | Fan, Q, Zhou, H, Li, X, Wang, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.05 Å) | | Cite: | Precise Control Over Kinetics of Molecular Assembly: Production of Particles with Tunable Sizes and Crystalline Forms.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8G92
 
 | | Structure of inhibitor 16d-bound SPNS2 | | Descriptor: | 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Chen, H, Li, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and functional insights into Spns2-mediated transport of sphingosine-1-phosphate.
Cell, 186, 2023
|
|
7VX2
 
 | | Crystal Structure of the Y53F/N55A/I80F/L114V/I116V mutant of LEH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2021-11-12 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.485 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
7VWD
 
 | | Crystal Structure of the Y53F/N55A mutant of LEH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2021-11-10 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
7VWM
 
 | | Crystal Structure of the Y53F/N55A/I116V mutant of LEH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2021-11-11 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
8G94
 
 | | Structure of CD69-bound S1PR1 coupled to heterotrimeric Gi | | Descriptor: | Early activation antigen CD69, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Chen, H, Li, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-19 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Transmembrane protein CD69 acts as an S1PR1 agonist.
Elife, 12, 2023
|
|
7XLT
 
 | | Cryo-EM Structure of R-loop monoclonal antibody S9.6 in recognizing RNA:DNA hybrids | | Descriptor: | DNA, RNA, S9.6 Fab HC, ... | | Authors: | Li, Q, Lin, C, Luo, Z, Li, H, Li, X, Sun, Q. | | Deposit date: | 2022-04-22 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of R-loop monoclonal antibody S9.6 in recognizing RNA:DNA hybrids.
J Genet Genomics, 49, 2022
|
|
8IHQ
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 | | Descriptor: | Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8J85
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 mutant S88E in complex with ochratoxin A | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-04-30 | | Release date: | 2023-08-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8IHS
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with ochratoxin A | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8IHR
 
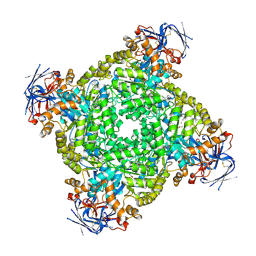 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with Phe | | Descriptor: | Amidohydrolase family protein, PHENYLALANINE, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8YN8
 
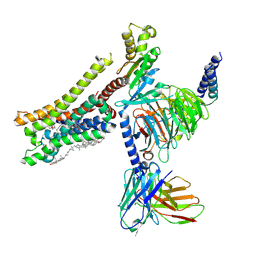 | | Cryo-EM structure of histamine H3 receptor in complex with proxyfan and miniGo | | Descriptor: | 5-(3-phenylmethoxypropyl)-1H-imidazole, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
8YN4
 
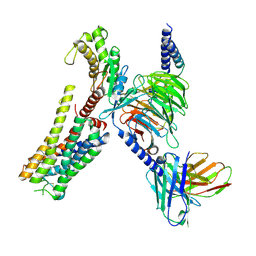 | | Cryo-EM structure of histamine H2 receptor in complex with histamine and miniGq | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Engineered guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
