6CCV
 
 | | Crystal structure of a Mycobacterium smegmatis RNA polymerase transcription initiation complex with inhibitor Rifampicin | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Rifamycin congeners kanglemycins are active against rifampicin-resistant bacteria via a distinct mechanism.
Nat Commun, 9, 2018
|
|
2RR4
 
 | | Complex structure of the zf-CW domain and the H3K4me3 peptide | | Descriptor: | Histone H3, ZINC ION, Zinc finger CW-type PWWP domain protein 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-03-24 | | Release date: | 2010-09-15 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the zinc finger CW domain as a histone modification reader
Structure, 18, 2010
|
|
6DCF
 
 | | Crystal structure of a Mycobacterium smegmatis transcription initiation complex with Rifampicin-resistant RNA polymerase and bound to kanglemycin A | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Rifamycin congeners kanglemycins are active against rifampicin-resistant bacteria via a distinct mechanism.
Nat Commun, 9, 2018
|
|
2CU7
 
 | | Solution structure of the SANT domain of human KIAA1915 protein | | Descriptor: | KIAA1915 protein | | Authors: | Yoneyama, M, Umehara, T, Saito, K, Tochio, N, Koshiba, S, Inoue, M, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-25 | | Release date: | 2005-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Differences of SWIRM Domain Subtypes
J.Mol.Biol., 369, 2007
|
|
3ARN
 
 | | Human dUTPase in complex with novel uracil derivative | | Descriptor: | Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION, N-{5-[(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)methoxy]-2-methylpentan-2-yl}benzenesulfonamide | | Authors: | Chong, K.T, Miyahara, S, Miyakoshi, H, Fukuoka, M. | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a novel class of potent human deoxyuridine triphosphatase inhibitors remarkably enhancing the antitumor activity of thymidylate synthase inhibitors
J.Med.Chem., 55, 2012
|
|
4ELC
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A C134 mutant with MTSEA modified Cys-165 | | Descriptor: | (2S)-2-hydroxybutanedioic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Botulinum neurotoxin A light chain, ... | | Authors: | Stura, E.A, Vera, L, Ptchelkine, D, Bakirci, H, Garcia, S, Dive, V. | | Deposit date: | 2012-04-10 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Framework for Covalent Inhibition of Clostridium botulinum Neurotoxin A by Targeting Cys165.
J.Biol.Chem., 287, 2012
|
|
4EL4
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A C134S/C165S double mutant | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin A light chain, GLYCEROL, ... | | Authors: | Stura, E.A, Vera, L, Ptchelkine, D, Dive, V. | | Deposit date: | 2012-04-10 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Framework for Covalent Inhibition of Clostridium botulinum Neurotoxin A by Targeting Cys165.
J.Biol.Chem., 287, 2012
|
|
7FHS
 
 | | Crystal structure of DYRK1A in complex with RD0392 | | Descriptor: | (5~{Z})-5-[(3-ethoxy-4-oxidanyl-phenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, GLYCEROL | | Authors: | Kikuchi, M, Sumida, T, Hosoya, T, Kii, I, Umehara, T. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure-activity relationship for the folding intermediate-selective inhibition of DYRK1A.
Eur.J.Med.Chem., 227, 2022
|
|
7FHT
 
 | | Crystal structure of DYRK1A in complex with RD0448 | | Descriptor: | (5~{Z})-5-[(3-ethynyl-4-methoxy-phenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Kikuchi, M, Sumida, Y, Hosoya, T, Kii, I, Umehara, T. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure-activity relationship for the folding intermediate-selective inhibition of DYRK1A.
Eur.J.Med.Chem., 227, 2022
|
|
4EJ5
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A wild-type | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin A light chain, CARBONATE ION, ... | | Authors: | Stura, E.A, Vera, L, Dive, V. | | Deposit date: | 2012-04-06 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Framework for Covalent Inhibition of Clostridium botulinum Neurotoxin A by Targeting Cys165.
J.Biol.Chem., 287, 2012
|
|
4HNO
 
 | | High resolution crystal structure of DNA Apurinic/apyrimidinic (AP) endonuclease IV Nfo from Thermatoga maritima | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Shin, D.S, Hosfield, D.J, Arvai, A.S, Tsutakawa, S.E, Tainer, J.A. | | Deposit date: | 2012-10-20 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.9194 Å) | | Cite: | Conserved Structural Chemistry for Incision Activity in Structurally Non-homologous Apurinic/Apyrimidinic Endonuclease APE1 and Endonuclease IV DNA Repair Enzymes.
J.Biol.Chem., 288, 2013
|
|
2RPJ
 
 | | Solution structure of Fn14 CRD domain | | Descriptor: | Tumor necrosis factor receptor superfamily member 12A | | Authors: | He, F, Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-05-19 | | Release date: | 2009-03-24 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cysteine-rich domain in Fn14, a member of the tumor necrosis factor receptor superfamily
Protein Sci., 18, 2009
|
|
7VMR
 
 | | Structure of recombinant RyR2 mutant K4593A (EGTA dataset) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMM
 
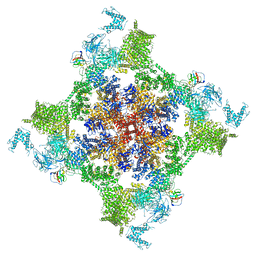 | | Structure of recombinant RyR2 (EGTA dataset, class 1, closed state) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMS
 
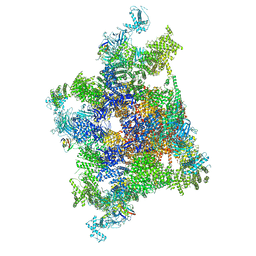 | | Structure of recombinant RyR2 mutant K4593A (Ca2+ dataset) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations
Nat Commun, 13, 2022
|
|
7VML
 
 | | Structure of recombinant RyR2 (EGTA dataset, class 1&2, closed state) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMN
 
 | | Structure of recombinant RyR2 (EGTA dataset, class 2, closed state) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMO
 
 | | Structure of recombinant RyR2 (Ca2+ dataset, class 1, open state) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMP
 
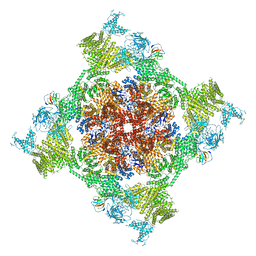 | | Structure of recombinant RyR2 (Ca2+ dataset, class 2, open state) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMQ
 
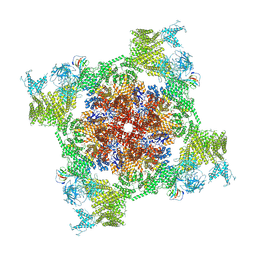 | | Structure of recombinant RyR2 (Ca2+ dataset, class 3, open state) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations
Nat Commun, 13, 2022
|
|
2YZ1
 
 | |
2YUE
 
 | | Solution structure of the NEUZ (NHR) domain in Neuralized from Drosophila melanogaster | | Descriptor: | Protein neuralized | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of the NHR1 domain of the Drosophila neuralized E3 ligase in the notch signaling pathway.
J.Mol.Biol., 393, 2009
|
|
3W07
 
 | |
1GP4
 
 | | Anthocyanidin synthase from Arabidopsis thaliana (selenomethionine substituted) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-OXOGLUTARIC ACID, ANTHOCYANIDIN SYNTHASE | | Authors: | Wilmouth, R.C, Turnbull, J.J, Welford, R.W.D, Clifton, I.J, Prescott, A.G, Schofield, C.J. | | Deposit date: | 2001-10-30 | | Release date: | 2002-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Mechanism of Anthocyanidin Synthase from Arabidopsis Thaliana.
Structure, 10, 2002
|
|
1GP6
 
 | | Anthocyanidin synthase from Arabidopsis thaliana complexed with trans-dihydroquercetin (with 30 min exposure to O2) | | Descriptor: | (2S,3S)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,5,7,3',4'-PENTAHYDROXYFLAVONE, ... | | Authors: | Wilmouth, R.C, Turnbull, J.J, Welford, R.W.D, Clifton, I.J, Prescott, A.G, Schofield, C.J. | | Deposit date: | 2001-10-30 | | Release date: | 2002-02-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Mechanism of Anthocyanidin Synthase from Arabidopsis Thaliana.
Structure, 10, 2002
|
|
