6FS1
 
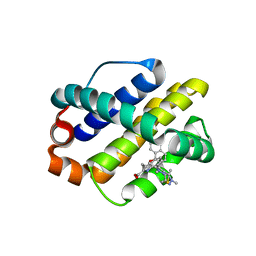 | | MCL1 in complex with an indole acid ligand | | Descriptor: | 1,2-ETHANEDIOL, 7-[3-[(1,5-dimethylpyrazol-3-yl)methylsulfanylmethyl]-1,5-dimethyl-pyrazol-4-yl]-3-(3-naphthalen-1-yloxypropyl)-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Kasmirski, S, Hargreaves, D. | | Deposit date: | 2018-02-18 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia.
Nat Commun, 9, 2018
|
|
8X83
 
 | |
8X82
 
 | |
8X84
 
 | |
6FS2
 
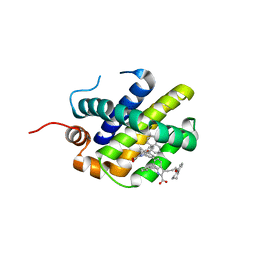 | | MCL1 in complex with indole acid ligand | | Descriptor: | 7-(2-methylphenyl)-3-[3-(5,6,7,8-tetrahydronaphthalen-1-yloxy)propyl]-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Hargreaves, D. | | Deposit date: | 2018-02-18 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia.
Nat Commun, 9, 2018
|
|
6FS0
 
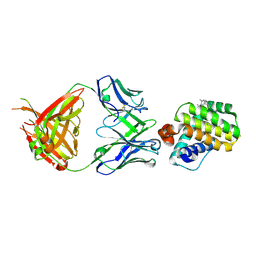 | |
3WD5
 
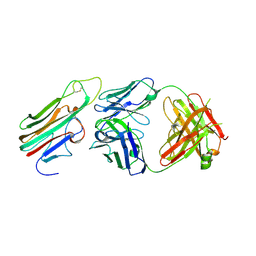 | | Crystal structure of TNFalpha in complex with Adalimumab Fab fragment | | Descriptor: | Adalimumab Heavy Chain, Adalimumab Light Chain, Tumor necrosis factor | | Authors: | Hu, S, Liang, S.Y, Guo, Y.J, Lou, Z.Y. | | Deposit date: | 2013-06-06 | | Release date: | 2013-08-14 | | Last modified: | 2015-07-01 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Comparison of the inhibition mechanisms of adalimumab and infliximab in treating tumor necrosis factor alpha-associated diseases from a molecular view
J.Biol.Chem., 288, 2013
|
|
7D3I
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with MI-23 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[3-[3,5-bis(fluoranyl)phenyl]propanoyl]-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase | | Authors: | Zeng, R, Li, Y.S, Qiao, J.X, Wang, Y.F, Yang, S.Y, Lei, J. | | Deposit date: | 2020-09-19 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
6INH
 
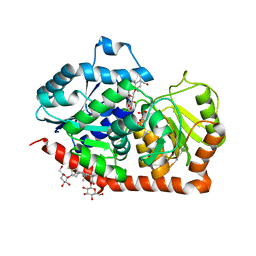 | | A glycosyltransferase with UDP and the substrate | | Descriptor: | 1-O-[(8alpha,9beta,10alpha,13alpha)-13-(beta-D-glucopyranosyloxy)-18-oxokaur-16-en-18-yl]-beta-D-glucopyranose, GLYCEROL, UDP-glycosyltransferase 76G1, ... | | Authors: | Zhu, X. | | Deposit date: | 2018-10-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
6ING
 
 | |
2M01
 
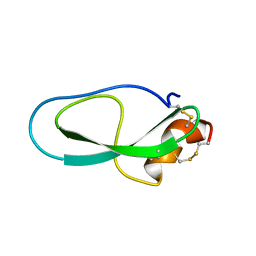 | | Solution structure of Kunitz-type neurotoxin LmKKT-1a from scorpion venom | | Descriptor: | Protease inhibitor LmKTT-1a | | Authors: | Luo, F, Jiang, L, Liu, M, Chen, Z, Wu, Y. | | Deposit date: | 2012-10-15 | | Release date: | 2013-11-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Genomic and structural characterization of Kunitz-type peptide LmKTT-1a highlights diversity and evolution of scorpion potassium channel toxins.
Plos One, 8, 2013
|
|
2UZQ
 
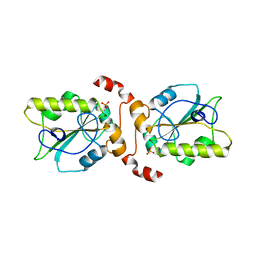 | | Protein Phosphatase, New Crystal Form | | Descriptor: | M-PHASE INDUCER PHOSPHATASE 2, PHOSPHATE ION | | Authors: | Hillig, R.C, Eberspaecher, U. | | Deposit date: | 2007-05-01 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | New Crystal Form of Protein Phosphatase Cdc25B Triggered by Guanidinium Chloride as an Additive
To be Published
|
|
3TYV
 
 | |
3DB6
 
 | | Crystal structure of an activated (Thr->Asp) Polo-like kinase 1 (Plk1) catalytic domain in complex with Compound 902 | | Descriptor: | 1-[5-methyl-2-(trifluoromethyl)furan-3-yl]-3-[(2Z)-5-(2-{[6-(1H-1,2,4-triazol-3-ylamino)pyrimidin-4-yl]amino}ethyl)-1,3-thiazol-2(3H)-ylidene]urea, Polo-like kinase 1 | | Authors: | Elling, R.A, Fucini, R.V, Zhu, J, Barr, K.J, Romanowski, M.J. | | Deposit date: | 2008-05-30 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of the Brachydanio rerio Polo-like kinase 1 (Plk1) catalytic domain in complex with an extended inhibitor targeting the adaptive pocket of the enzyme.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
3DXX
 
 | | Crystal structure of EcTrmB | | Descriptor: | GLYCEROL, tRNA (guanine-N(7)-)-methyltransferase | | Authors: | Zhou, H.H, Liu, Q, Gao, Y.X, Teng, M.K, Niu, L.W. | | Deposit date: | 2008-07-25 | | Release date: | 2009-05-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Monomeric tRNA (m(7)G46) methyltransferase from Escherichia coli presents a novel structure at the function-essential insertion
Proteins, 76, 2009
|
|
1B62
 
 | | MUTL COMPLEXED WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (MUTL) | | Authors: | Wei, Y. | | Deposit date: | 1999-01-11 | | Release date: | 1999-04-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair.
Cell(Cambridge,Mass.), 97, 1999
|
|
3DXZ
 
 | | Crystal structure of EcTrmB in complex with SAH | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Zhou, H.H, Liu, Q, Gao, Y.X, Teng, M.K, Niu, L.W. | | Deposit date: | 2008-07-25 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Monomeric tRNA (m(7)G46) methyltransferase from Escherichia coli presents a novel structure at the function-essential insertion
Proteins, 76, 2009
|
|
3DXY
 
 | | Crystal structure of EcTrmB in complex with SAM | | Descriptor: | GLYCEROL, S-ADENOSYLMETHIONINE, SULFATE ION, ... | | Authors: | Zhou, H.H, Liu, Q, Gao, Y.X, Teng, M.K, Niu, L.W. | | Deposit date: | 2008-07-25 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Monomeric tRNA (m(7)G46) methyltransferase from Escherichia coli presents a novel structure at the function-essential insertion
Proteins, 76, 2009
|
|
5DV2
 
 | |
5DV4
 
 | |
3HI5
 
 | | Crystal structure of Fab fragment of AL-57 | | Descriptor: | Heavy chain of Fab fragment of AL-57 against alpha L I domain, light chain of Fab fragment of AL-57 against alpha L I domain | | Authors: | Zhang, H, Wang, J. | | Deposit date: | 2009-05-18 | | Release date: | 2009-09-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of activation-dependent binding of ligand-mimetic antibody AL-57 to integrin LFA-1.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5X8I
 
 | | Crystal structure of human CLK1 in complex with compound 25 | | Descriptor: | 5-[1-[(1S)-1-(4-fluorophenyl)ethyl]-[1,2,3]triazolo[4,5-c]quinolin-8-yl]-1,3-benzoxazole, Dual specificity protein kinase CLK1 | | Authors: | Sun, Q.Z, Lin, G.F, Li, L.L, Jin, X.T, Huang, L.Y, Zhang, G, Wei, Y.Q, Lu, G.W, Yang, S.Y. | | Deposit date: | 2017-03-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Cdc2-Like Kinase 1 (CLK1) as a New Class of Autophagy Inducers
J. Med. Chem., 60, 2017
|
|
6NZM
 
 | | Brutons tyrosine kinase in complex with compound 50. | | Descriptor: | 1,2-ETHANEDIOL, N-[2-fluoro-6-(pyrrolidin-1-yl)phenyl]-N'-{3-[(2R)-1-(2-hydroxyethyl)-4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperazin-2-yl]phenyl}urea, Tyrosine-protein kinase BTK | | Authors: | Marcotte, D.J. | | Deposit date: | 2019-02-14 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Optimization of novel reversible Bruton's tyrosine kinase inhibitors identified using Tethering-fragment-based screens.
Bioorg.Med.Chem., 27, 2019
|
|
4Z62
 
 | | The plant peptide hormone free receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Phytosulfokine receptor 1 | | Authors: | Chai, J, Wang, J, Han, Z. | | Deposit date: | 2015-04-03 | | Release date: | 2016-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric receptor activation by the plant peptide hormone phytosulfokine
Nature, 525, 2015
|
|
4Z5W
 
 | | The plant peptide hormone receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Phytosulfokine, ... | | Authors: | Chai, J, Wang, J, Han, Z. | | Deposit date: | 2015-04-03 | | Release date: | 2016-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Allosteric receptor activation by the plant peptide hormone phytosulfokine
Nature, 525, 2015
|
|
