7MYO
 
 | | Cryo-EM structure of p110alpha in complex with p85alpha inhibited by BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | Deposit date: | 2021-05-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3JCS
 
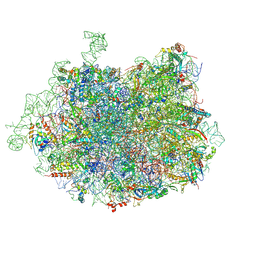 | | 2.8 Angstrom cryo-EM structure of the large ribosomal subunit from the eukaryotic parasite Leishmania | | Descriptor: | 26S alpha ribosomal RNA, 26S delta ribosomal RNA, 26S epsilon ribosomal RNA, ... | | Authors: | Shalev-Benami, M, Zhang, Y, Matzov, D, Halfon, Y, Zackay, A, Rozenberg, H, Zimmerman, E, Bashan, A, Jaffe, C.L, Yonath, A, Skiniotis, G. | | Deposit date: | 2016-01-21 | | Release date: | 2016-07-20 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | 2.8- angstrom Cryo-EM Structure of the Large Ribosomal Subunit from the Eukaryotic Parasite Leishmania.
Cell Rep, 16, 2016
|
|
3JA8
 
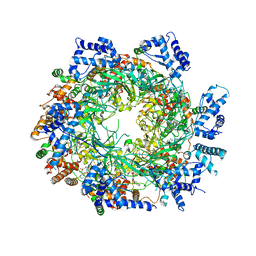 | | Cryo-EM structure of the MCM2-7 double hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Minichromosome Maintenance 2, Minichromosome Maintenance 3, ... | | Authors: | Li, N, Zhai, Y, Zhang, Y, Li, W, Yang, M, Lei, J, Tye, B.K, Gao, N. | | Deposit date: | 2015-05-09 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the eukaryotic MCM complex at 3.8 angstrom
Nature, 524, 2015
|
|
2LD7
 
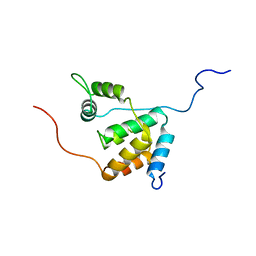 | | Solution structure of the mSin3A PAH3-SAP30 SID complex | | Descriptor: | Histone deacetylase complex subunit SAP30, Paired amphipathic helix protein Sin3a | | Authors: | Xie, T, He, Y, Korkeamaki, H, Zhang, Y, Imhoff, R, Lohi, O, Radhakrishnan, I. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the 30-kDa Sin3-associated protein (SAP30) in complex with the mammalian Sin3A corepressor and its role in nucleic acid binding.
J.Biol.Chem., 286, 2011
|
|
3JSF
 
 | |
3JSG
 
 | |
3JTU
 
 | |
8H02
 
 | |
4HX1
 
 | | Structure of HLA-A68 complexed with a tumor antigen derived peptide | | Descriptor: | 9-mer peptide from Tyrosinase-related protein-2, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Niu, L, Cheng, H, Zhang, S, Tan, S, Zhang, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2012-11-09 | | Release date: | 2013-10-02 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural basis for the differential classification of HLA-A*6802 and HLA-A*6801 into the A2 and A3 supertypes
Mol.Immunol., 55, 2013
|
|
4I48
 
 | | Structure of HLA-A68 complexed with an HIV Env derived peptide | | Descriptor: | 9-mer peptide from Envelope glycoprotein gp160, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Niu, L, Cheng, H, Zhang, S, Tan, S, Zhang, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2012-11-27 | | Release date: | 2013-10-02 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structural basis for the differential classification of HLA-A*6802 and HLA-A*6801 into the A2 and A3 supertypes
Mol.Immunol., 55, 2013
|
|
4HWZ
 
 | | Structure of HLA-A68 complexed with an HIV derived peptide | | Descriptor: | 9-mer peptide from Pol protein, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Niu, L, Cheng, H, Zhang, S, Tan, S, Zhang, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2012-11-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structural basis for the differential classification of HLA-A*6802 and HLA-A*6801 into the A2 and A3 supertypes
Mol.Immunol., 55, 2013
|
|
3ORW
 
 | | Crystal structure of thermophilic phosphotriesterase from Geobacillus kaustophilus HTA426 | | Descriptor: | COBALT (II) ION, Phosphotriesterase | | Authors: | Zheng, B.S, Yu, S.S, Zhang, Y, Lou, Z.Y, Feng, Y. | | Deposit date: | 2010-09-07 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of thermophilic phosphotriesterase from Geobacillus kaustophilus HTA426
To be Published
|
|
3L5R
 
 | |
3L5U
 
 | |
3L5S
 
 | |
3L5V
 
 | |
3L5P
 
 | |
5I2D
 
 | | Crystal structure of T. thermophilus TTHB099 class II transcription activation complex: TAP-RPo | | Descriptor: | DNA (72-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Feng, Y, Zhang, Y, Ebright, R.H. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.405 Å) | | Cite: | Structural basis of transcription activation.
Science, 352, 2016
|
|
1L3K
 
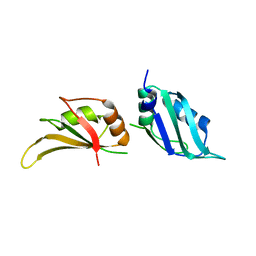 | | UP1, THE TWO RNA-RECOGNITION MOTIF DOMAIN OF HNRNP A1 | | Descriptor: | HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN A1 | | Authors: | Vitali, J, Ding, J, Jiang, J, Zhang, Y, Krainer, A.R, Xu, R.-M. | | Deposit date: | 2002-02-27 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Correlated alternative side chain conformations in the RNA-recognition motif of heterogeneous nuclear ribonucleoprotein A1.
Nucleic Acids Res., 30, 2002
|
|
1K4V
 
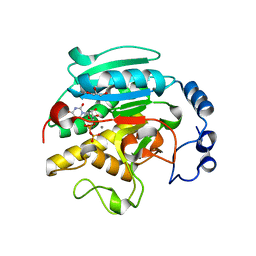 | | 1.53 A Crystal Structure of the Beta-Galactoside-alpha-1,3-galactosyltransferase in Complex with UDP | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Boix, E, Swaminathan, G.J, Zhang, Y, Natesh, R, Brew, K, Acharya, K.R. | | Deposit date: | 2001-10-09 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure of UDP complex of UDP-galactose:beta-galactoside-alpha -1,3-galactosyltransferase at 1.53-A resolution reveals a conformational change in the catalytically important C terminus.
J.Biol.Chem., 276, 2001
|
|
5HN7
 
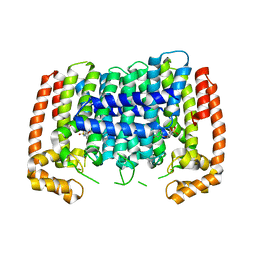 | |
5HNA
 
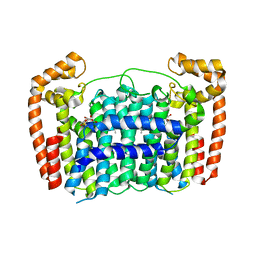 | |
5HN8
 
 | | Crystal structure of Plasmodium vivax geranylgeranylpyrophosphate synthase complexed with BPH-1182 | | Descriptor: | 2-{[3-hydroxy-5-(octyloxy)benzyl]sulfanyl}benzoic acid, Farnesyl pyrophosphate synthase, putative, ... | | Authors: | Liu, Y.-L, Zhang, Y, Oldfield, E. | | Deposit date: | 2016-01-18 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dynamic Structure and Inhibition of a Malaria Drug Target: Geranylgeranyl Diphosphate Synthase.
Biochemistry, 55, 2016
|
|
5HN9
 
 | |
8H9I
 
 | | Human ATP synthase F1 domain, state2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit O, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
