5JA7
 
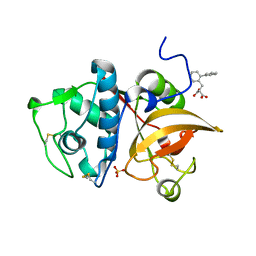 | | Human cathepsin K mutant C25S in complex with the allosteric effector NSC94914 | | Descriptor: | ACETATE ION, Cathepsin K, GLYCEROL, ... | | Authors: | Novinec, M, Korenc, M, Lenarcic, B. | | Deposit date: | 2016-04-12 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | An allosteric site enables fine-tuning of cathepsin K by diverse effectors.
FEBS Lett., 590, 2016
|
|
9BHI
 
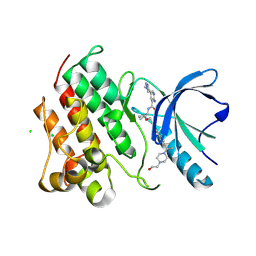 | | Crystal structure of the MerTK kinase domain with SA4488 | | Descriptor: | (5P)-2-amino-5-(1-methyl-1H-pyrazol-4-yl)-N-{(1R,2S)-2-[(4'-{2-[4-(2-oxoethyl)piperazin-1-yl]propan-2-yl}[1,1'-biphenyl]-4-yl)methoxy]cyclopentyl}pyridine-3-carboxamide, CHLORIDE ION, Mer tyrosine kinase domain | | Authors: | Jakob, C.G, Qui, W, Jain, R. | | Deposit date: | 2024-04-20 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of A-910, a Highly Potent and Orally Bioavailable Dual MerTK/Axl-Selective Tyrosine Kinase Inhibitor.
J.Med.Chem., 67, 2024
|
|
6ZCF
 
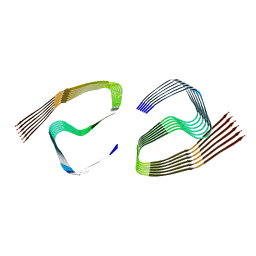 | |
6ZCG
 
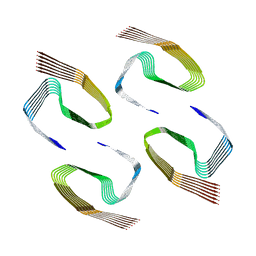 | |
6ZCH
 
 | |
8RMT
 
 | | Galectin-3 with a bound inhibitor | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-~{N}-[3,5-bis(chloranyl)phenyl]-6-(hydroxymethyl)-3-methoxy-5-oxidanyl-~{N}-[(1~{S},2~{S})-2-oxidanylcyclohexyl]-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-2-carboxamide, Galectin-3, THIOCYANATE ION | | Authors: | Mac Sweeney, A, Sager, C. | | Deposit date: | 2024-01-08 | | Release date: | 2025-01-29 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of Galactopyranose-1-carboxamides as a New Class of Small, Novel, Potent, Selective, and Orally Active Galectin-3 Inhibitors.
Chemmedchem, 20, 2025
|
|
8RMV
 
 | | Galectin-3 with a bound inhibitor | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-~{N}-[3,5-bis(chloranyl)phenyl]-6-(hydroxymethyl)-3-methoxy-5-oxidanyl-~{N}-[(1~{S},2~{S})-2-oxidanylcyclobutyl]-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-2-carboxamide, Galectin-3, THIOCYANATE ION | | Authors: | Mac Sweeney, A, Sager, C. | | Deposit date: | 2024-01-08 | | Release date: | 2025-01-29 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery of Galactopyranose-1-carboxamides as a New Class of Small, Novel, Potent, Selective, and Orally Active Galectin-3 Inhibitors.
Chemmedchem, 20, 2025
|
|
8RMU
 
 | | Galectin-3 with a bound inhibitor | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-~{N}-[3,5-bis(chloranyl)phenyl]-6-(hydroxymethyl)-3-methoxy-5-oxidanyl-~{N}-[(1~{S},2~{S})-2-oxidanylcyclopentyl]-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-2-carboxamide, Galectin-3, THIOCYANATE ION | | Authors: | Mac Sweeney, A, Sager, C. | | Deposit date: | 2024-01-08 | | Release date: | 2025-01-29 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery of Galactopyranose-1-carboxamides as a New Class of Small, Novel, Potent, Selective, and Orally Active Galectin-3 Inhibitors.
Chemmedchem, 20, 2025
|
|
6FKK
 
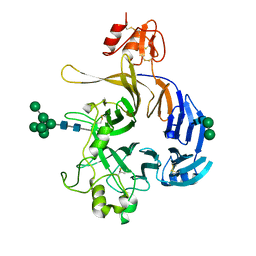 | | Drosophila Semaphorin 1b, extracellular domains 1-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MIP07328p, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rozbesky, D, Harlos, K, Jones, E.Y. | | Deposit date: | 2018-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Diversity of oligomerization in Drosophila semaphorins suggests a mechanism of functional fine-tuning.
Nat Commun, 10, 2019
|
|
8DGX
 
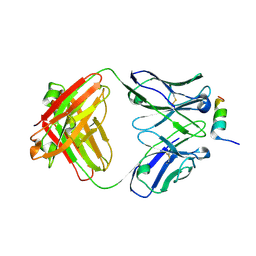 | |
8DGW
 
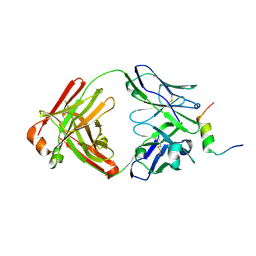 | |
8DGU
 
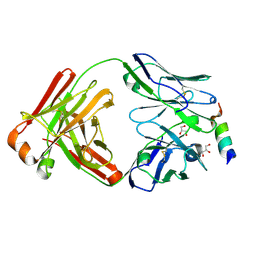 | |
8DGV
 
 | |
8PXC
 
 | | Structure of Fap1, a domain of the accessory Sec-dependent serine-rich glycoprotein adhesin from Streptococcus oralis, solved at wavelength 3.06 A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Fap1 | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Owen, C.D, Walsh, M.A, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
6GG0
 
 | |
6GXY
 
 | | Tryparedoxin from Trypanosoma brucei in complex with CFT | | Descriptor: | 5-(4-fluorophenyl)-2-methyl-3~{H}-thieno[2,3-d]pyrimidin-4-one, Tryparedoxin | | Authors: | Bader, N, Wagner, A, Hellmich, U, Schindelin, H. | | Deposit date: | 2018-06-27 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitor-Induced Dimerization of an Essential Oxidoreductase from African Trypanosomes.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
7B27
 
 | |
4LP7
 
 | |
5LXK
 
 | | NMR structure of the C-terminal domain of the Bacteriophage T5 decoration protein pb10. | | Descriptor: | Decoration protein | | Authors: | Vernhes, E, Gilquin, B, Cuniasse, P, Boulanger, P, Zinn-Justin, S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid.
Sci Rep, 7, 2017
|
|
6GVW
 
 | | Crystal structure of the BRCA1-A complex | | Descriptor: | BRCA1-A complex subunit Abraxas 1, BRCA1-A complex subunit RAP80, BRISC and BRCA1-A complex member 1, ... | | Authors: | Bunker, R.D, Rabl, J, Thoma, N.H. | | Deposit date: | 2018-06-21 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Structural Basis of BRCC36 Function in DNA Repair and Immune Regulation.
Mol.Cell, 75, 2019
|
|
6GXG
 
 | | Tryparedoxin from Trypanosoma brucei in complex with CFT | | Descriptor: | 5-(4-fluorophenyl)-2-methyl-3~{H}-thieno[2,3-d]pyrimidin-4-one, GLYCEROL, Tryparedoxin | | Authors: | Bader, N, Wagner, A, Hellmich, U, Schindelin, H. | | Deposit date: | 2018-06-27 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibitor-Induced Dimerization of an Essential Oxidoreductase from African Trypanosomes.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6H3C
 
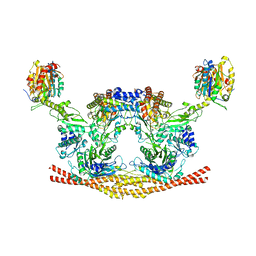 | | Cryo-EM structure of the BRISC complex bound to SHMT2 | | Descriptor: | BRISC and BRCA1-A complex member 1, BRISC and BRCA1-A complex member 2, BRISC complex subunit Abraxas 2, ... | | Authors: | Bunker, R.D, Rabl, J, Thoma, N.H. | | Deposit date: | 2018-07-18 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis of BRCC36 Function in DNA Repair and Immune Regulation.
Mol.Cell, 75, 2019
|
|
5LXL
 
 | | NMR structure of the N-terminal domain of the Bacteriophage T5 decoration protein pb10 | | Descriptor: | Decoration protein | | Authors: | Vernhes, E, Gilquin, B, Cuniasse, P, Boulanger, P, Zinn-Justin, S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid.
Sci Rep, 7, 2017
|
|
8PYV
 
 | | Structure of Human PS-1 GSH-analog complex, solved at wavelength 2.755 A | | Descriptor: | L-gamma-glutamyl-S-(2-biphenyl-4-yl-2-oxoethyl)-L-cysteinylglycine, PALMITIC ACID, Prostaglandin E synthase | | Authors: | Duman, R, El Omari, K, Mykhaylyk, V, Orr, C, Wagner, A, Vogeley, L, Brown, D.G. | | Deposit date: | 2023-07-26 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX9
 
 | | Structure of the antibacterial peptide ABC transporter McjD, solved at wavelength 2.75 A | | Descriptor: | MAGNESIUM ION, Microcin-J25 export ATP-binding/permease protein McjD, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Bountra, K, Beis, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
