6KJ2
 
 | | 200kV MicroED structure of FUS (37-42) SYSGYS solved from single crystal at 0.67 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.67 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
4RFP
 
 | |
6KG7
 
 | | Cryo-EM Structure of the Mammalian Tactile Channel Piezo2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Piezo-type mechanosensitive ion channel component 2 | | Authors: | Wang, L, Zhou, H, Zhang, M, Liu, W, Deng, T, Zhao, Q, Li, Y, Lei, J, Li, X, Xiao, B. | | Deposit date: | 2019-07-11 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and mechanogating of the mammalian tactile channel PIEZO2.
Nature, 573, 2019
|
|
6KJ1
 
 | | 200kV MicroED structure of FUS (37-42) SYSGYS solved from merged datasets at 0.65 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.65 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
6KY4
 
 | | Crystal structure of Sulfiredoxin from Arabidopsis thaliana | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, Sulfiredoxin, ... | | Authors: | Liu, M, Wang, J, Li, X, Li, M, Sylvanno, M.J, Zhang, M, Wang, M. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of sulfiredoxin from Arabidopsis thaliana revealed a more robust antioxidant mechanism in plants.
Biochem.Biophys.Res.Commun., 520, 2019
|
|
4QY1
 
 | | Structure of H10 from human-infecting H10N8 in complex with avian receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
6IZO
 
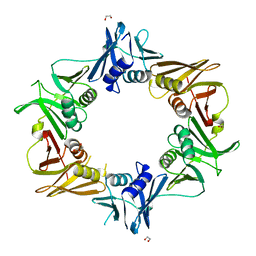 | | Crystal structure of DNA polymerase sliding clamp from Caulobacter crescentus | | Descriptor: | 1,2-ETHANEDIOL, Beta sliding clamp, DI(HYDROXYETHYL)ETHER | | Authors: | Jiang, X, Zhang, L, Teng, M, Li, X. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Caulobacter crescentus beta sliding clamp employs a noncanonical regulatory model of DNA replication.
Febs J., 287, 2020
|
|
4IFS
 
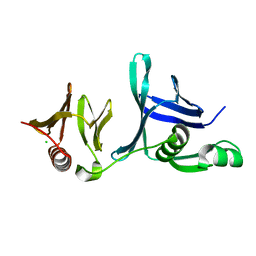 | | Crystal structure of the hSSRP1 Middle domain | | Descriptor: | CHLORIDE ION, FACT complex subunit SSRP1 | | Authors: | Zhang, W.J, Zeng, F.X, Shao, C, Liu, Y.W, Niu, L.W, Li, X, Teng, M.K. | | Deposit date: | 2012-12-15 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of the hSSRP1 Middle domain
To be Published
|
|
5ZV7
 
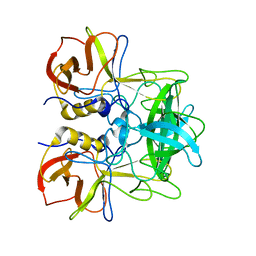 | | P domain of GII.17-2014/15 complexed with B-trisaccharide | | Descriptor: | VP1, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5ZVC
 
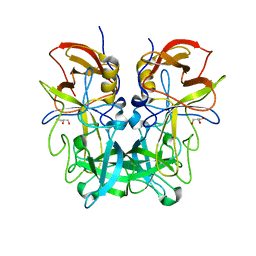 | | P domain of GII.13 norovirus capsid complexed with Lewis A trisaccharide | | Descriptor: | GLYCEROL, Major capsid protein VP1, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-10 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5ZV9
 
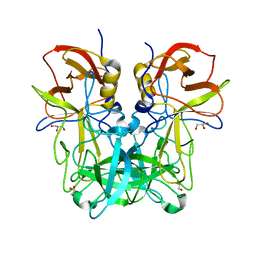 | | P domain of GII.13 norovirus capsid | | Descriptor: | GLYCEROL, Major capsid protein VP1 | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
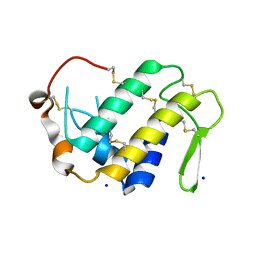
4H0S
 
 | |
5WJ9
 
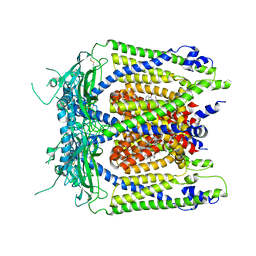 | | Human TRPML1 channel structure in agonist-bound open conformation | | Descriptor: | 2-{2-oxo-2-[(4S)-2,2,4-trimethyl-3,4-dihydroquinolin-1(2H)-yl]ethyl}-1H-isoindole-1,3(2H)-dione, Mucolipin-1 | | Authors: | Schmiege, P, Li, X. | | Deposit date: | 2017-07-21 | | Release date: | 2017-10-18 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Human TRPML1 channel structures in open and closed conformations.
Nature, 550, 2017
|
|
6JIR
 
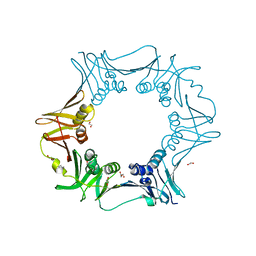 | | Crystal structure of C. crescentus beta sliding clamp with PEG bound to putative beta-motif tethering region | | Descriptor: | 1,2-ETHANEDIOL, Beta sliding clamp, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jiang, X, Teng, M, Li, X. | | Deposit date: | 2019-02-23 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Caulobacter crescentus beta sliding clamp employs a noncanonical regulatory model of DNA replication.
Febs J., 287, 2020
|
|
4HCT
 
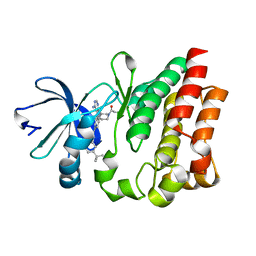 | | Crystal structure of ITK in complex with compound 52 | | Descriptor: | 3-{1-[(3R)-1-acryloylpiperidin-3-yl]-4-amino-1H-pyrazolo[3,4-d]pyrimidin-3-yl}-N-(3-tert-butylphenyl)benzamide, Tyrosine-protein kinase ITK/TSK | | Authors: | Zapf, C.W, Gerstenberger, B.S, Xing, L, Limburg, D.C, Anderson, D.R, Caspers, N, Han, S, Aulabaugh, A, Kurumbail, R, Shakya, S, Li, X, Spaulding, V, Czerwinski, R.M, Seth, N, Medley, Q.G. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Covalent inhibitors of interleukin-2 inducible T cell kinase (itk) with nanomolar potency in a whole-blood assay.
J.Med.Chem., 55, 2012
|
|
5ZV5
 
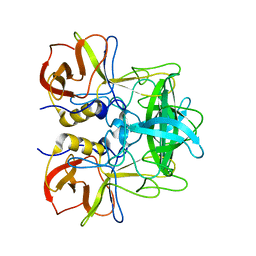 | | P domain of GII.17-2014/15 complexed with A-trisaccharide | | Descriptor: | VP1, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5WQ7
 
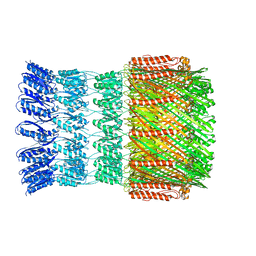 | |
7URC
 
 | | Human PORCN in complex with LGK974 | | Descriptor: | 2-[(2P)-2',3-dimethyl[2,4'-bipyridin]-5-yl]-N-[(5P)-5-(pyrazin-2-yl)pyridin-2-yl]acetamide, 2C11 heavy chain, 2C11 light chain, ... | | Authors: | Liu, Y, Qi, X, Li, X. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-20 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Mechanisms and inhibition of Porcupine-mediated Wnt acylation.
Nature, 607, 2022
|
|
7URD
 
 | | Human PORCN in complex with LGK974 and WNT3A peptide | | Descriptor: | 2-[(2P)-2',3-dimethyl[2,4'-bipyridin]-5-yl]-N-[(5P)-5-(pyrazin-2-yl)pyridin-2-yl]acetamide, 2C11 heavy chain, 2C11 light chain, ... | | Authors: | Liu, Y, Qi, X, Li, X. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-20 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Mechanisms and inhibition of Porcupine-mediated Wnt acylation.
Nature, 607, 2022
|
|
8YAG
 
 | | Cryo-electron microscopic structure of an amide hydrolase from Pseudoxanthomonas wuyuanensis | | Descriptor: | Imidazolonepropionase, ZINC ION | | Authors: | Dai, L.H, Xu, Y.H, Hu, Y.M, Niu, D, Yang, X.C, Shen, P.P, Li, X, Xie, Z.Z, Li, H, Guo, R.-T, Chen, C.-C. | | Deposit date: | 2024-02-09 | | Release date: | 2024-10-09 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Functional characterization and structural basis of an efficient ochratoxin A-degrading amidohydrolase.
Int.J.Biol.Macromol., 278, 2024
|
|
4IQ8
 
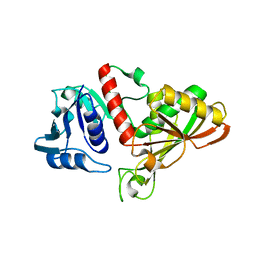 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 3 from Saccharomyces cerevisiae | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase 3 | | Authors: | Wang, H, Liu, Q, Niu, L, Teng, M, Li, X. | | Deposit date: | 2013-01-11 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Preliminary crystallographic analysis of glyceraldehyde-3-phosphate dehydrogenase 3 from Saccharomyces cerevisiae.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4J1J
 
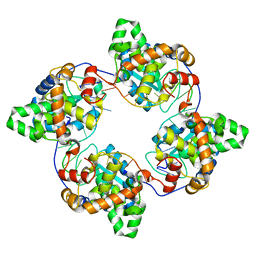 | | Leanyer orthobunyavirus nucleoprotein-ssDNA complex | | Descriptor: | DNA (5'-D(P*AP*CP*CP*AP*AP*AP*CP*AP*AP*CP*CP*CP*AP*CP*CP*CP*A)-3'), Nucleocapsid | | Authors: | Niu, F, Shaw, N, Wang, Y, Jiao, L, Ding, W, Li, X, Zhu, P, Upur, H, Ouyang, S, Cheng, G, Liu, Z.J. | | Deposit date: | 2013-02-01 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the Leanyer orthobunyavirus nucleoprotein-RNA complex reveals unique architecture for RNA encapsidation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5WQ9
 
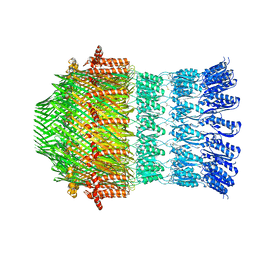 | |
5WQ8
 
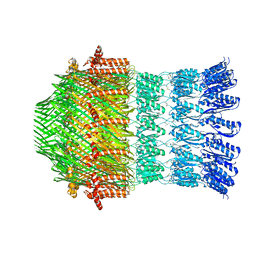 | |
5V93
 
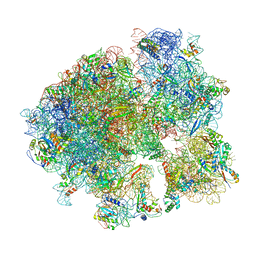 | | Cryo-EM structure of the 70S ribosome from Mycobacterium tuberculosis bound with Capreomycin | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Yang, K, Chang, J.-Y, Cui, Z, Li, X, Meng, R, Duan, L, Thongchol, J, Jakana, J, Huwe, C, Sacchettini, J, Zhang, J. | | Deposit date: | 2017-03-22 | | Release date: | 2017-09-20 | | Last modified: | 2020-08-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the human pathogen Mycobacterium tuberculosis.
Nucleic Acids Res., 45, 2017
|
|
