6DRX
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5HT2B receptor, BRIL chimera, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DS0
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (1S)-1-[(2-chloro-3,4-dimethoxyphenyl)methyl]-6-methyl-2,3,4,9-tetrahydro-1H-beta-carboline, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5HT2B receptor, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.188 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1RF2
 
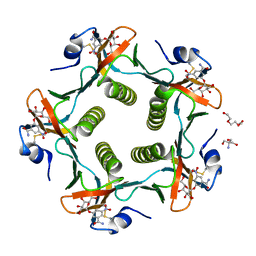 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV4 | | Descriptor: | 1,3-BIS-([3-[3-[3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYLAMINO-3,4-DIOXO-CYCLOBUTENYL]-AMINO-PROPOXY-ETHOXY-ETHOXY]-PROPYL-]AMINO-CARBONYLOXY)-2-AMINO-PROPANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TRIETHYLENE GLYCOL, ... | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-07 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
5AJM
 
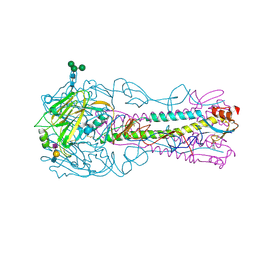 | | H5 (VN1194) Asn186Lys Mutant Haemagglutinin in Complex with Avian Receptor Analogue 3'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Xiong, X, Xiao, H, Martin, S.R, Coombs, P.J, Liu, J, Collins, P.J, Vachieri, S.G, Walker, P.A, Lin, Y.P, McCauley, J.W, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Enhanced Human Receptor Binding by H5 Haemagglutinins.
Virology, 456, 2014
|
|
1RD9
 
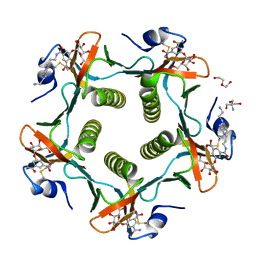 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV2 | | Descriptor: | 1,3-BIS-([3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYL-AMINO]-CARBONYLOXY)-2-AMINO-PROPANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HEXAETHYLENE GLYCOL, ... | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-05 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
3EN2
 
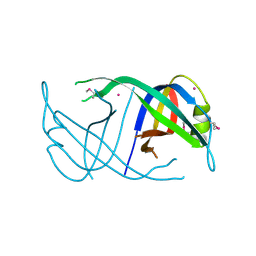 | | Three-dimensional structure of the protein priB from Ralstonia solanacearum at the resolution 2.3A. Northeast Structural Genomics Consortium target RsR213C. | | Descriptor: | POTASSIUM ION, Probable primosomal replication protein n | | Authors: | Kuzin, A.P, Neely, H, Wang, H, Sahdev, S, Foote, E.L, Xiao, R, Liu, J, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the protein priB from Ralstonia solanacearum at the resolution 2.3A. Northeast Structural Genomics Consortium target RsR213C.
To be Published
|
|
1R28
 
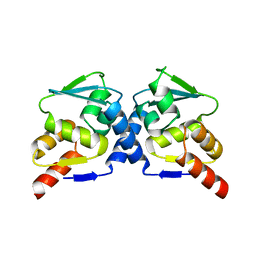 | | Crystal Structure of the B-Cell Lymphoma 6 (BCL6) BTB domain to 2.2 Angstrom | | Descriptor: | B-cell lymphoma 6 protein | | Authors: | Ahmad, K.F, Melnick, A, Lax, S.A, Bouchard, D, Liu, J, Kiang, C.L, Mayer, S, Licht, J.D, Prive, G.G. | | Deposit date: | 2003-09-26 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain.
Mol.Cell, 12, 2003
|
|
3ELO
 
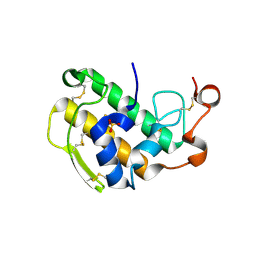 | | Crystal Structure of Human Pancreatic Prophospholipase A2 | | Descriptor: | Phospholipase A2, SULFATE ION | | Authors: | Xu, W, Yi, L, Feng, Y, Chen, L, Liu, J. | | Deposit date: | 2008-09-22 | | Release date: | 2009-04-14 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insight into the activation mechanism of human pancreatic prophospholipase A2
J.Biol.Chem., 284, 2009
|
|
1RDP
 
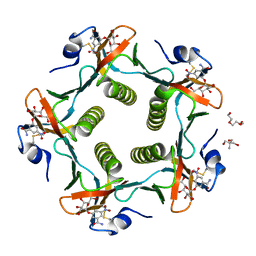 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV3 | | Descriptor: | 1,3-BIS-([[3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYLAMINO-3,4-DIOXO-CYCLOBU TENYL]-AMINO-ETHYL]-AMINO-CARBONYLOXY)-2-AMINO-PROPANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TRIETHYLENE GLYCOL, ... | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-05 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
4EJQ
 
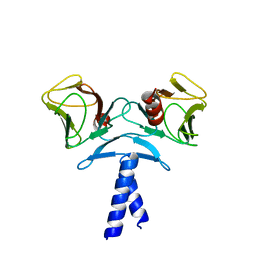 | | Crystal structure of KIF1A C-CC1-FHA | | Descriptor: | Kinesin-like protein KIF1A | | Authors: | Huo, L, Yue, Y, Ren, J, Yu, J, Liu, J, Yu, Y, Ye, F, Xu, T, Zhang, M, Feng, W. | | Deposit date: | 2012-04-06 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | The CC1-FHA Tandem as a Central Hub for Controlling the Dimerization and Activation of Kinesin-3 KIF1A
Structure, 20, 2012
|
|
3D3E
 
 | | Crystal Structure of Human 11-beta-Hydroxysteroid Dehydrogenase (HSD1) in Complex with Benzamide Inhibitor | | Descriptor: | Corticosteroid 11-beta-dehydrogenase isozyme 1, N-cyclopropyl-N-(trans-4-pyridin-3-ylcyclohexyl)-4-[(1S)-2,2,2-trifluoro-1-hydroxy-1-methylethyl]benzamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, Z, Sudom, A, Liu, J, Walker, N.P. | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Novel, Potent Benzamide Inhibitors of 11beta-Hydroxysteroid Dehydrogenase Type 1 (11beta-HSD1) Exhibiting Oral Activity in an Enzyme Inhibition ex Vivo Model
J.Med.Chem., 51, 2008
|
|
4LLA
 
 | | Crystal structure of D3D4 domain of the LILRB2 molecule | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily B member 2 | | Authors: | Nam, G, Shi, Y, Ryu, M, Wang, Q, Song, H, Liu, J, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-09 | | Release date: | 2013-09-11 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal structures of the two membrane-proximal Ig-like domains (D3D4) of LILRB1/B2: alternative models for their involvement in peptide-HLA binding
Protein Cell, 4, 2013
|
|
4LL9
 
 | | Crystal structure of D3D4 domain of the LILRB1 molecule | | Descriptor: | IODIDE ION, Leukocyte immunoglobulin-like receptor subfamily B member 1 | | Authors: | Nam, G, Shi, Y, Ryu, M, Wang, Q, Song, H, Liu, J, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-09 | | Release date: | 2013-09-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | Crystal structures of the two membrane-proximal Ig-like domains (D3D4) of LILRB1/B2: alternative models for their involvement in peptide-HLA binding
Protein Cell, 4, 2013
|
|
1T8T
 
 | | Crystal Structure of human 3-O-Sulfotransferase-3 with bound PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, CITRIC ACID, heparan sulfate D-glucosaminyl 3-O-sulfotransferase 3A1 | | Authors: | Moon, A.F, Edavettal, S.C, Krahn, J.M, Munoz, E.M, Negishi, M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2004-05-13 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of the sulfotransferase (3-o-sulfotransferase isoform 3) involved in the biosynthesis of an entry receptor for herpes simplex virus 1
J.Biol.Chem., 279, 2004
|
|
1RCV
 
 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV1 | | Descriptor: | [3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYLAMINO] -2-(3-{4-[3-(3-NITRO-5-[GALACTOPYRANOSYLOXY]-BENZOYLAMINO)-PROPYL]-PIPERAZIN-1-YL} -PROPYL-AMINO)-3,4-DIOXO-CYCLOBUTENE, cholera toxin B protein (CTB) | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-04 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
1T8U
 
 | | Crystal Structure of human 3-O-Sulfotransferase-3 with bound PAP and tetrasaccharide substrate | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ADENOSINE-3'-5'-DIPHOSPHATE, SODIUM ION, ... | | Authors: | Moon, A.F, Edavettal, S.C, Krahn, J.M, Munoz, E.M, Negishi, M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2004-05-13 | | Release date: | 2004-08-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of the sulfotransferase (3-o-sulfotransferase isoform 3) involved in the biosynthesis of an entry receptor for herpes simplex virus 1
J.Biol.Chem., 279, 2004
|
|
1TU4
 
 | | Crystal Structure of Rab5-GDP Complex | | Descriptor: | COBALT (II) ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-5A, ... | | Authors: | Zhu, G, Zhai, P, Liu, J, Terzyan, S, Li, G, Zhang, X.C. | | Deposit date: | 2004-06-24 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of Rab5-Rabaptin5 interaction in endocytosis
Nat.Struct.Mol.Biol., 11, 2004
|
|
5TTF
 
 | | Crystal structure of catalytic domain of G9a with MS012 | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EHMT2, N4-(1-methylpiperidin-4-yl)-N2-hexyl-6,7-dimethoxyquinazoline-2,4-diamine, ... | | Authors: | DONG, A, ZENG, H, LIU, J, XIONG, Y, BABAULT, N, JIN, J, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, WU, H, BROWN, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-03 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase.
J. Med. Chem., 60, 2017
|
|
5TUZ
 
 | | Structure of human GLP SET-domain (EHMT1) in complex with inhibitor MS0124 | | Descriptor: | 1,2-ETHANEDIOL, 6,7-dimethoxy-N-(1-methylpiperidin-4-yl)-2-(morpholin-4-yl)quinazolin-4-amine, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2016-11-07 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase.
J. Med. Chem., 60, 2017
|
|
4HX1
 
 | | Structure of HLA-A68 complexed with a tumor antigen derived peptide | | Descriptor: | 9-mer peptide from Tyrosinase-related protein-2, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Niu, L, Cheng, H, Zhang, S, Tan, S, Zhang, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2012-11-09 | | Release date: | 2013-10-02 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural basis for the differential classification of HLA-A*6802 and HLA-A*6801 into the A2 and A3 supertypes
Mol.Immunol., 55, 2013
|
|
6LKE
 
 | | in meso full-length rat KMO in complex with an inhibitor identified via DNA-encoded chemical library screening | | Descriptor: | 4-chloranyl-2-[[5-chloranyl-2-(5-methoxy-1,3-dihydroisoindol-2-yl)-1,3-thiazol-4-yl]carbonyl-methyl-amino]-5-fluoranyl-benzoic acid, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mimasu, S, Yamagishi, H, Kiyohara, M, Hupp, D.C, Liu, J, Kakefuda, K, Okuda, T. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Full-length in meso structure and mechanism of rat kynurenine 3-monooxygenase inhibition.
Commun Biol, 4, 2021
|
|
4IKB
 
 | | Crystal structure of SNX11 PX domain | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Xu, J, Xu, T, Liu, J. | | Deposit date: | 2012-12-26 | | Release date: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of Sorting Nexin 11 (SNX11) Reveals a Novel Extended PX Domain (PXe Domain) Critical for the Inhibition of Sorting Nexin 10 (SNX10) Induced Vacuolation
to be published
|
|
5GW8
 
 | | Crystal structure of a putative DAG-like lipase (MgMDL2) from Malassezia globosa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Xu, J, Xu, H, Liu, J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Malassezia globosa MgMDL2 lipase: Crystal structure and rational modification of substrate specificity.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
4FAO
 
 | | Specificity and Structure of a high affinity Activin-like 1 (ALK1) signaling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2B, Growth/differentiation factor 2, ... | | Authors: | Townson, S.A, Martinez-Hackert, E, Greppi, C, Lowden, P, Sako, D, Liu, J, Ucran, J.A, Liharska, K, Underwood, K.W, Seehra, J, Kumar, R, Grinberg, A.V. | | Deposit date: | 2012-05-22 | | Release date: | 2012-06-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.357 Å) | | Cite: | Specificity and Structure of a High Affinity Activin Receptor-like Kinase 1 (ALK1) Signaling Complex.
J.Biol.Chem., 287, 2012
|
|
4IKD
 
 | | Crystal structure of SNX11 PX domain | | Descriptor: | CHLORIDE ION, SODIUM ION, Sorting nexin-11 | | Authors: | Xu, J, Xu, T, Liu, J. | | Deposit date: | 2012-12-26 | | Release date: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Sorting Nexin 11 (SNX11) Reveals a Novel Extended PX Domain (PXe Domain) Critical for the Inhibition of Sorting Nexin 10 (SNX10) Induced Vacuolation
to be published
|
|
