8A2P
 
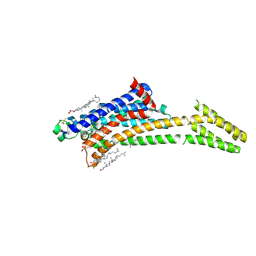 | | Room-temperature structure of the stabilised A2A-LUAA47070 complex determined by synchrotron serial crystallography | | Descriptor: | 4-(3,3-dimethylbutanoylamino)-3,5-bis(fluoranyl)-~{N}-(1,3-thiazol-2-yl)benzamide, Adenosine receptor A2a,Soluble cytochrome b562, CHOLESTEROL, ... | | Authors: | Moraes, I, Kwan, T.O.C, Axford, D. | | Deposit date: | 2022-06-06 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A versatile approach to high-density microcrystals in lipidic cubic phase for room-temperature serial crystallography.
J.Appl.Crystallogr., 56, 2023
|
|
8A2O
 
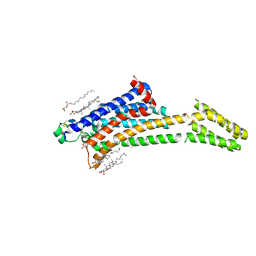 | | Room-temperature structure of the stabilised A2A-Theophylline complex determined by synchrotron serial crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562, CHOLESTEROL, ... | | Authors: | Moraes, I, Kwan, T.O.C, Axford, D. | | Deposit date: | 2022-06-06 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | A versatile approach to high-density microcrystals in lipidic cubic phase for room-temperature serial crystallography.
J.Appl.Crystallogr., 56, 2023
|
|
6CXZ
 
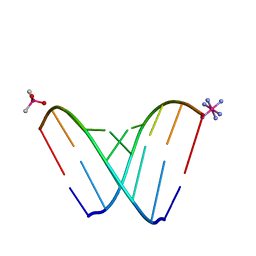 | | RNA octamer containing 2'-F, 4'-Calpha-Me U. | | Descriptor: | CACODYLATE ION, COBALT HEXAMMINE(III), RNA (5'-R(*CP*GP*AP*AP*(U4M)P*UP*CP*G)-3') | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2018-04-04 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the synergy of 4'- and 2'-modifications on siRNA nuclease resistance, thermal stability and RNAi activity.
Nucleic Acids Res., 46, 2018
|
|
1LWB
 
 | |
470D
 
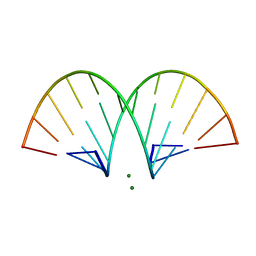 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
468D
 
 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
1RMS
 
 | | CRYSTAL STRUCTURES OF RIBONUCLEASE MS COMPLEXED WITH 3'-GUANYLIC ACID A GP*C ANALOGUE, 2'-DEOXY-2'-FLUOROGUANYLYL-3',5'-CYTIDINE | | Descriptor: | GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE MS | | Authors: | Nonaka, T, Mitsui, Y, Nakamura, K.T. | | Deposit date: | 1991-12-02 | | Release date: | 1992-07-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ribonuclease Ms (as a ribonuclease T1 homologue) complexed with a guanylyl-3',5'-cytidine analogue.
Biochemistry, 32, 1993
|
|
2G92
 
 | |
1RDS
 
 | | CRYSTAL STRUCTURE OF RIBONUCLEASE MS (AS RIBONUCLEASE T1 HOMOLOGUE) COMPLEXED WITH A GUANYLYL-3',5'-CYTIDINE ANALOGUE | | Descriptor: | 2'-FLUOROGUANYLYL-(3'-5')-PHOSPHOCYTIDINE, RIBONUCLEASE MS | | Authors: | Nonaka, T, Nakamura, K.T, Mitsui, Y. | | Deposit date: | 1993-05-14 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of ribonuclease Ms (as a ribonuclease T1 homologue) complexed with a guanylyl-3',5'-cytidine analogue.
Biochemistry, 32, 1993
|
|
2FII
 
 | |
2FIJ
 
 | |
1R77
 
 | | Crystal structure of the cell wall targeting domain of peptidylglycan hydrolase ALE-1 | | Descriptor: | Cell Wall Targeting Domain of Glycylglycine Endopeptidase ALE-1 | | Authors: | Lu, J.Z, Fujiwara, T, Komatsuzawa, H, Sugai, M, Sakon, J. | | Deposit date: | 2003-10-20 | | Release date: | 2005-04-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cell Wall-targeting Domain of Glycylglycine Endopeptidase Distinguishes among Peptidoglycan Cross-bridges.
J.Biol.Chem., 281, 2006
|
|
5YDN
 
 | | Mu pahge neck subunit | | Descriptor: | Gene product J | | Authors: | Takeda, S, Iwasaki, T, Yamashita, E, Nakagawa, A. | | Deposit date: | 2017-09-13 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three-dimensional structures of bacteriophage neck subunits are shared in Podoviridae, Siphoviridae and Myoviridae
Genes Cells, 23, 2018
|
|
2FIL
 
 | |
5WRN
 
 | |
2FIH
 
 | |
1UB3
 
 | | Crystal Structure of Tetrameric Structure of Aldolase from thermus thermophilus HB8 | | Descriptor: | 1-HYDROXY-PENTANE-3,4-DIOL-5-PHOSPHATE, Aldolase protein | | Authors: | Lokanath, N.K, Miyano, M, Yokoyama, S, Kuramitsu, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-03-28 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of aldolase from Thermus thermophilus HB8 showing the contribution of oligomeric state to thermostability.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1J2W
 
 | | Tetrameric Structure of aldolase from Thermus thermophilus HB8 | | Descriptor: | Aldolase protein | | Authors: | Lokanath, N.K, Shiromizu, I, Miyano, M, Yokoyama, S, Kuramitsu, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-01-14 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of aldolase from Thermus thermophilus HB8 showing the contribution of oligomeric state to thermostability.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1I20
 
 | | MUTANT HUMAN LYSOZYME (A92D) | | Descriptor: | LYSOZYME C | | Authors: | Kuroki, R. | | Deposit date: | 2001-02-05 | | Release date: | 2001-02-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic responses of mutations at a Ca2+ binding site engineered into human lysozyme.
J.Biol.Chem., 273, 1998
|
|
1I1Z
 
 | | MUTANT HUMAN LYSOZYME (Q86D) | | Descriptor: | LYSOZYME C | | Authors: | Kuroki, R. | | Deposit date: | 2001-02-05 | | Release date: | 2001-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic responses of mutations at a Ca2+ binding site engineered into human lysozyme.
J.Biol.Chem., 273, 1998
|
|
1I22
 
 | | MUTANT HUMAN LYSOZYME (A83K/Q86D/A92D) | | Descriptor: | CALCIUM ION, LYSOZYME C | | Authors: | Kuroki, R. | | Deposit date: | 2001-02-05 | | Release date: | 2001-02-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic responses of mutations at a Ca2+ binding site engineered into human lysozyme.
J.Biol.Chem., 273, 1998
|
|
4DZB
 
 | |
1KP4
 
 | |
1WR8
 
 | |
1WNL
 
 | |
