6P47
 
 | | Cryo-EM structure of TMEM16F in digitonin without calcium | | Descriptor: | Anoctamin-6 | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
7RMH
 
 | | Substance P bound to active human neurokinin 1 receptor in complex with miniGs399 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Harris, J.A, Faust, B, Gondin, A.B, Daemgen, M.A, Suomivuori, C.M, Veldhuis, N.A, Cheng, Y, Dror, R.O, Thal, D, Manglik, A. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-03 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Selective G protein signaling driven by substance P-neurokinin receptor dynamics.
Nat.Chem.Biol., 18, 2022
|
|
8JUN
 
 | | Cryo-EM structure of SIDT1 E555Q mutant | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SID1 transmembrane family member 1, ZINC ION | | Authors: | Sun, C.R, Xu, D, Li, Q, Zhou, C.Z, Chen, Y. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Human SIDT1 mediates dsRNA uptake via its phospholipase activity.
Cell Res., 34, 2024
|
|
8JUL
 
 | | Cryo-EM structure of SIDT1 in complex with phosphatidic acid | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, SID1 transmembrane family member 1, ZINC ION | | Authors: | Sun, C.R, Xu, D, Li, Q, Zhou, C.Z, Chen, Y. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Human SIDT1 mediates dsRNA uptake via its phospholipase activity.
Cell Res., 34, 2024
|
|
3S8G
 
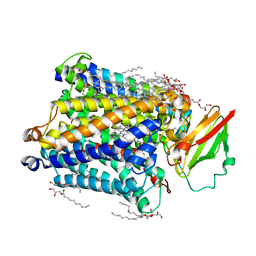 | | 1.8 A structure of ba3 cytochrome c oxidase mutant (A120F) from Thermus thermophilus in lipid environment | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Tiefenbrunn, T, Liu, W, Chen, Y, Katritch, V, Stout, C.D, Fee, J.A, Cherezov, V. | | Deposit date: | 2011-05-27 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment.
Plos One, 6, 2011
|
|
3S8F
 
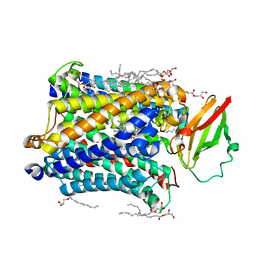 | | 1.8 A structure of ba3 cytochrome c oxidase from Thermus thermophilus in lipid environment | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Tiefenbrunn, T, Liu, W, Chen, Y, Katritch, V, Stout, C.D, Fee, J.A, Cherezov, V. | | Deposit date: | 2011-05-27 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment.
Plos One, 6, 2011
|
|
6P48
 
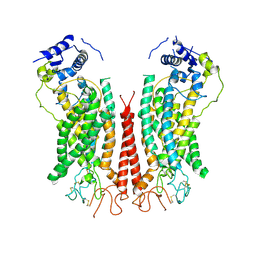 | | Cryo-EM structure of calcium-bound TMEM16F in nanodisc with supplement of PIP2 in Cl1 | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
6VHS
 
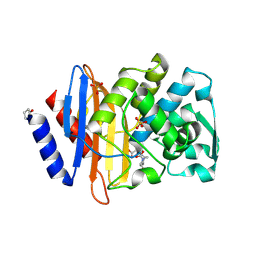 | | Crystal structure of CTX-M-14 in complex with beta-lactamase inhibitor ETX1317 | | Descriptor: | (2R)-({[(3R,6S)-6-carbamoyl-1-formyl-4-methyl-1,2,3,6-tetrahydropyridin-3-yl]amino}oxy)(fluoro)acetic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Sacco, M.D, Chen, Y. | | Deposit date: | 2020-01-10 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Discovery of an Orally Available Diazabicyclooctane Inhibitor (ETX0282) of Class A, C, and D Serine beta-Lactamases.
J.Med.Chem., 63, 2020
|
|
3TKL
 
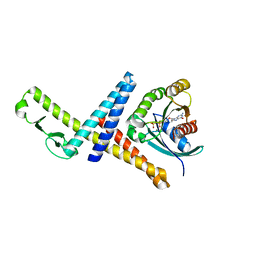 | | Crystal structure of the GTP-bound Rab1a in complex with the coiled-coil domain of LidA from Legionella pneumophila | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, LidA protein, substrate of the Dot/Icm system, ... | | Authors: | Cheng, W, Yin, K, Lu, D, Li, B, Zhu, D, Chen, Y, Zhang, H, Xu, S, Chai, J, Gu, L. | | Deposit date: | 2011-08-27 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.183 Å) | | Cite: | Structural insights into a unique Legionella pneumophila effector LidA recognizing both GDP and GTP bound Rab1 in their active state
Plos Pathog., 8, 2012
|
|
7U9B
 
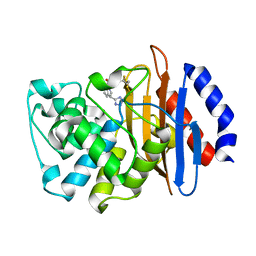 | | Crystal structure of indoline 7 with KPC-2 | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC, [(3S)-3-(1H-tetrazol-5-yl)-2,3-dihydro-1H-indol-1-yl][3-(trifluoromethyl)phenyl]methanone | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the binding hot spots of KPC-2 carbapenemase using reversible tetrazole-based inhibitors
To Be Published
|
|
8JSJ
 
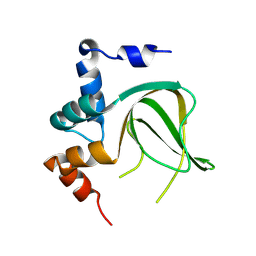 | |
8JSK
 
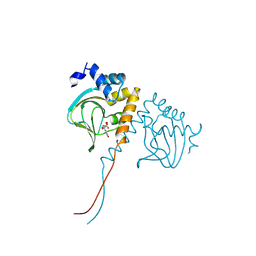 | | Crystal structure of an N-terminal cyclic nucleotide-binding domain of a PycTIR from Pseudovibrio sp. in complex with cUMP | | Descriptor: | PycTIR, Uridine-3',5'-cyclic monophosphate | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-06-20 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural and functional characterization of cyclic pyrimidine-regulated anti-phage system.
Nat Commun, 15, 2024
|
|
8JSZ
 
 | | Crystal structure of a uridylate cyclase from Anabaena sp. | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-06-20 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural and functional characterization of cyclic pyrimidine-regulated anti-phage system.
Nat Commun, 15, 2024
|
|
8JSF
 
 | |
7UA7
 
 | | Crystal structure of indoline 9 with KPC-2 | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, tert-butyl {(3R)-3-[(1H-tetrazol-5-yl)carbamoyl]-1-[3-(trifluoromethyl)benzoyl]-2,3-dihydro-1H-indol-3-yl}carbamate | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2022-03-11 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Probing the binding hot spots of KPC-2 carbapenemase using reversible tetrazole-based inhibitors
To Be Published
|
|
8I5E
 
 | | Crystal structure of HLA-A*11:01 in complex with KRAS peptide (VVGAGGVGK) | | Descriptor: | Beta-2-microglobulin, KRAS-G12wt-9, MHC class I antigen (Fragment) | | Authors: | Lu, D, Chen, Y, Jiang, M, Tan, S.G, Chai, Y, Gao, G.F. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a TCR in complex with HLA-A*11:01 bound to KRAS peptide (VVGAVGVGK)
Nat Commun, 2023
|
|
7MZ5
 
 | |
7MZ6
 
 | | Cryo-EM structure of minimal TRPV1 with 1 perturbed PI | | Descriptor: | (10R,13S)-16-amino-13-hydroxy-7,13-dioxo-8,12,14-trioxa-13lambda~5~-phosphahexadecan-10-yl hexadecanoate, (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZE
 
 | | Cryo-EM structure of minimal TRPV1 with 2 bound RTX in opposite pockets | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZB
 
 | | Cryo-EM structure of minimal TRPV1 with 3 bound RTX and 1 perturbed PI | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZC
 
 | |
7MZA
 
 | | Cryo-EM structure of minimal TRPV1 with 2 bound RTX in adjacent pockets | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZ9
 
 | | Cryo-EM structure of minimal TRPV1 with 1 partially bound RTX | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZ7
 
 | |
7MZD
 
 | |
