7ESZ
 
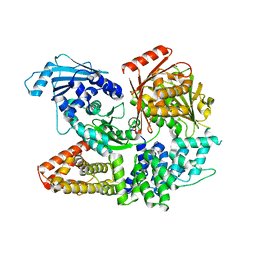 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CinA and CinB with Mn2+ from wPip | | Descriptor: | BACTERIA FACTOR A, BACTERIA FACTOR B, MANGANESE (II) ION | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.476 Å) | | Cite: | Structural and mechanistic insights into the complexes formed by Wolbachia cytoplasmic incompatibility factors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ET0
 
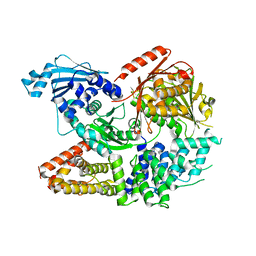 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CinA and CinB from wPip | | Descriptor: | Bacteria factor A, Bacteria factor B | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mechanistic insights into the complexes formed by Wolbachia cytoplasmic incompatibility factors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3C7K
 
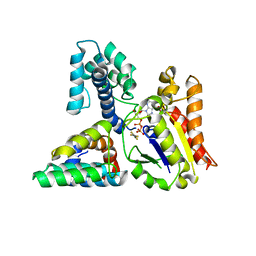 | | Molecular architecture of Galphao and the structural basis for RGS16-mediated deactivation | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(o) subunit alpha, MAGNESIUM ION, ... | | Authors: | Slep, K.C, Kercher, M.A, Wieland, T, Chen, C, Simon, M.I, Sigler, P.B. | | Deposit date: | 2008-02-07 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular architecture of G{alpha}o and the structural basis for RGS16-mediated deactivation.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3C7L
 
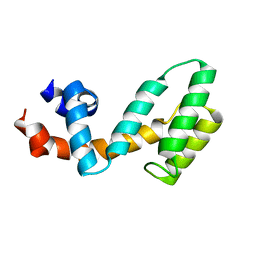 | | Molecular architecture of Galphao and the structural basis for RGS16-mediated deactivation | | Descriptor: | Regulator of G-protein signaling 16 | | Authors: | Slep, K.C, Kercher, M.A, Wieland, T, Chen, C, Simon, M.I, Sigler, P.B. | | Deposit date: | 2008-02-07 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular architecture of G{alpha}o and the structural basis for RGS16-mediated deactivation.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6OMO
 
 | | Human BMP6 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone morphogenetic protein 6, ... | | Authors: | Juo, Z.S, Seeherman, H. | | Deposit date: | 2019-04-19 | | Release date: | 2019-05-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A BMP/activin A chimera is superior to native BMPs and induces bone repair in nonhuman primates when delivered in a composite matrix.
Sci Transl Med, 11, 2019
|
|
6OMN
 
 | | Glycosylated BMP2 homodimer | | Descriptor: | Bone morphogenetic protein 2, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Juo, Z.S, Seeherman, H. | | Deposit date: | 2019-04-19 | | Release date: | 2019-05-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | A BMP/activin A chimera is superior to native BMPs and induces bone repair in nonhuman primates when delivered in a composite matrix.
Sci Transl Med, 11, 2019
|
|
6OML
 
 | | Human BMP chimera BV261 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bone morphogenetic protein 2 and Bone morphogenetic protein 6, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Seeherman, H, Juo, Z.S. | | Deposit date: | 2019-04-18 | | Release date: | 2019-05-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A BMP/activin A chimera is superior to native BMPs and induces bone repair in nonhuman primates when delivered in a composite matrix.
Sci Transl Med, 11, 2019
|
|
2JRT
 
 | | NMR solution structure of the protein coded by gene RHOS4_12090 of Rhodobacter sphaeroides. Northeast Structural Genomics target RhR5 | | Descriptor: | Uncharacterized protein | | Authors: | Wang, L, Chen, C, Nwosu, C, Cunningham, K, Owens, L, Ma, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-28 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the protein coded by gene RHOS4_12090 of Rhodobacter sphaeroides.
To be Published
|
|
5TFM
 
 | | Crystal Structure of Human Cadherin-23 EC6-8 | | Descriptor: | CALCIUM ION, Cadherin-23, SODIUM ION | | Authors: | Jaiganesh, A, Sotomayor, M. | | Deposit date: | 2016-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
5TFK
 
 | |
5TFL
 
 | | Crystal Structure of Mouse Cadherin-23 EC7+8 | | Descriptor: | CALCIUM ION, Cadherin-23, SODIUM ION | | Authors: | Jaiganesh, A, Sotomayor, M. | | Deposit date: | 2016-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
5GNK
 
 | | Crystal structure of EGFR 696-988 T790M in complex with LXX-6-34 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3R)-3-[4-azanyl-3-[3-chloranyl-4-[(1-methylimidazol-2-yl)methoxy]phenyl]pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]prop-2-en-1-one, CHLORIDE ION, ... | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2016-07-21 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Discovery of (R)-1-(3-(4-Amino-3-(3-chloro-4-(pyridin-2-ylmethoxy)phenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl)piperidin-1-yl)prop-2-en-1-one (CHMFL-EGFR-202) as a Novel Irreversible EGFR Mutant Kinase Inhibitor with a Distinct Binding Mode.
J. Med. Chem., 60, 2017
|
|
5GTY
 
 | | Crystal structure of EGFR 696-1022 T790M in complex with LXX-6-26 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3R)-3-[4-azanyl-3-[3-chloranyl-4-[(6-methylpyridin-2-yl)methoxy]phenyl]pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]prop-2-en-1-one, Epidermal growth factor receptor | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Discovery and characterization of a novel irreversible EGFR mutants selective and potent kinase inhibitor CHMFL-EGFR-26 with a distinct binding mode.
Oncotarget, 8, 2017
|
|
7BZV
 
 | |
2DUA
 
 | |
7V2Z
 
 | | ZIKV NS3helicase in complex with ssRNA and ATP-Mn2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Core protein, MANGANESE (II) ION, ... | | Authors: | Lin, M.M, Yang, H.T. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.10101676 Å) | | Cite: | Structural Basis of Zika Virus Helicase in RNA Unwinding and ATP Hydrolysis.
Acs Infect Dis., 8, 2022
|
|
7BWQ
 
 | | Structure of nonstructural protein Nsp9 from SARS-CoV-2 | | Descriptor: | Nsp9, SULFATE ION | | Authors: | Zhang, C, Chen, Y, Li, L, Su, D. | | Deposit date: | 2020-04-15 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural basis for the multimerization of nonstructural protein nsp9 from SARS-CoV-2.
Mol Biomed, 1, 2020
|
|
6W4Q
 
 | | Crystal structure of full-length tailspike protein 2 (TSP2, ORF211) ) from Escherichia coli O157:H7 bacteriophage CAB120 | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Greenfield, J, Herzberg, O. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of bacteriophage CBA120 ORF211 (TSP2), the determinant of phage specificity towards E. coli O157:H7.
Sci Rep, 10, 2020
|
|
5J39
 
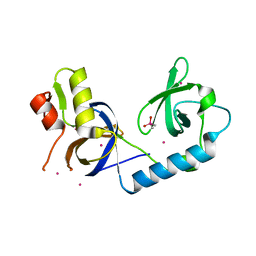 | | Crystal Structure of the extended TUDOR domain from TDRD2 | | Descriptor: | CACODYLATE ION, Tudor and KH domain-containing protein, UNKNOWN ATOM OR ION | | Authors: | Zhang, H, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-30 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for arginine methylation-independent recognition of PIWIL1 by TDRD2.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3HG8
 
 | |
8EQN
 
 | |
3IUR
 
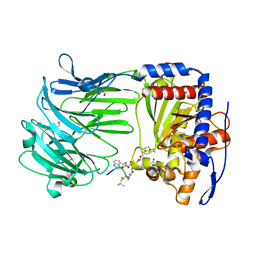 | | apPEP_D266Nx+H2H3 opened state | | Descriptor: | GLYCEROL, H2H3 helices from villin headpiece subdomain HP35, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
3IVM
 
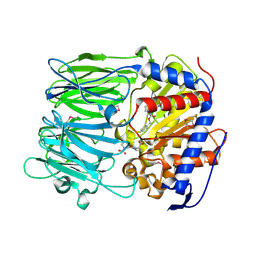 | | apPEP_WT+PP closed state | | Descriptor: | N-BENZYLOXYCARBONYL-L-PROLYL-L-PROLINAL, prolyl endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-09-01 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
3IUQ
 
 | | apPEP_D622N+PP closed state | | Descriptor: | GLYCEROL, N-BENZYLOXYCARBONYL-L-PROLYL-L-PROLINAL, Prolyl Endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
3IJK
 
 | | 5-OMe modified DNA 8mer | | Descriptor: | 5'-D(*GP*(UMS)P*GP*(T5O)P*AP*CP*AP*C)-3' | | Authors: | Sheng, J, Zhang, W, Hassan, A.E.A, Gan, J, Huang, Z. | | Deposit date: | 2009-08-04 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Synthesis of Pyrimidine Modified Seleno-DNA as a Novel Approach to Antisense Candidate
Chemistryselect, 8, 2023
|
|
