7M8K
 
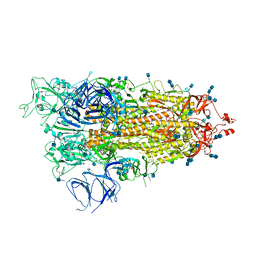 | | Cryo-EM structure of Brazil (P.1) SARS-CoV-2 spike glycoprotein variant in the prefusion state (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Casner, R.G, Cerutti, G, Shapiro, L, Ho, D.D. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-05 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY | | Cite: | Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization.
Cell Host Microbe, 29, 2021
|
|
8SQO
 
 | |
8SQR
 
 | |
4O3F
 
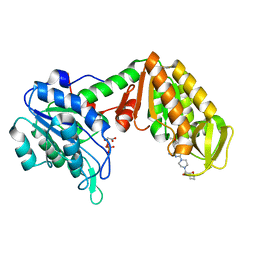 | | Crystal Structure of mouse PGK1 3PG and terazosin(TZN) ternary complex | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Phosphoglycerate kinase 1, [4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl][(2R)-tetrahydrofuran-2-yl]methanone | | Authors: | Li, X.L, Finci, I.L, Wang, J.H. | | Deposit date: | 2013-12-18 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Terazosin activates Pgk1 and Hsp90 to promote stress resistance.
Nat.Chem.Biol., 11, 2015
|
|
8DXS
 
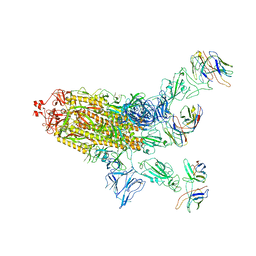 | |
8DW9
 
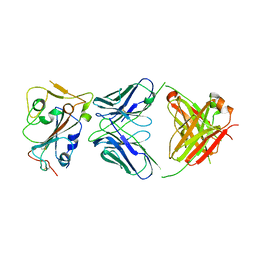 | |
8DWA
 
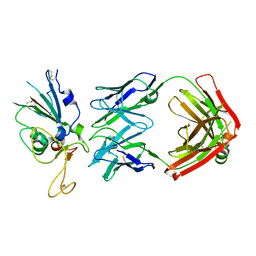 | |
8T7W
 
 | |
8SWD
 
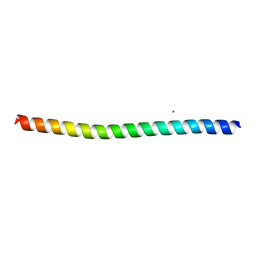 | |
8SQQ
 
 | |
8SQP
 
 | |
8SQT
 
 | |
8SU6
 
 | |
8T7Z
 
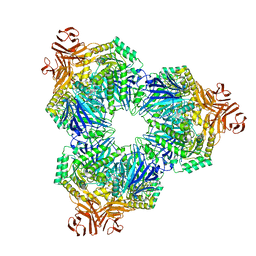 | |
5D7I
 
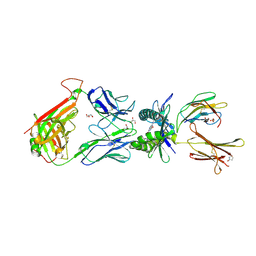 | | Structure of human MR1-Ac-6-FP in complex with human MAIT M33.64 TCR | | Descriptor: | Beta-2-microglobulin, GLYCEROL, M33.64 TCR Alpha Chain, ... | | Authors: | Keller, A.N, Woolley, R.E, Rossjohn, J. | | Deposit date: | 2015-08-14 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diversity of T Cells Restricted by the MHC Class I-Related Molecule MR1 Facilitates Differential Antigen Recognition.
Immunity, 44, 2016
|
|
5D5M
 
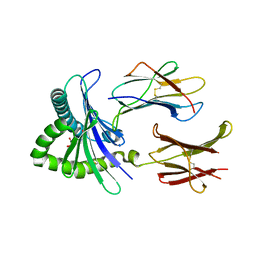 | | Structure of human MR1-5-OP-RU in complex with human MAIT M33.64 TCR | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Keller, A.N, Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2015-08-11 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Diversity of T Cells Restricted by the MHC Class I-Related Molecule MR1 Facilitates Differential Antigen Recognition.
Immunity, 44, 2016
|
|
7LBQ
 
 | |
7LBK
 
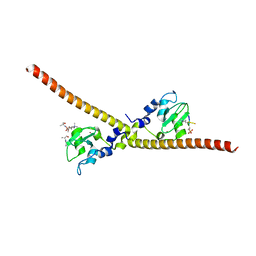 | |
7LBP
 
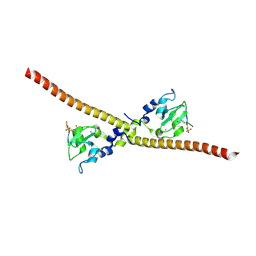 | |
7LBO
 
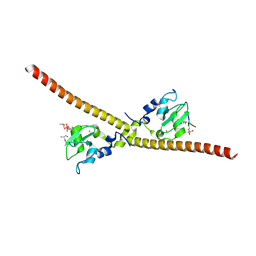 | |
6L42
 
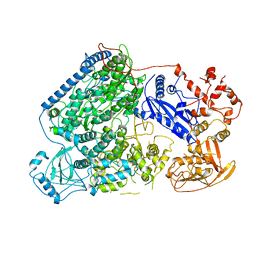 | |
6U25
 
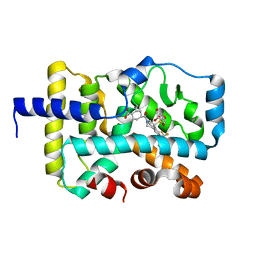 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C (NHIS- RORGT(244-487)-L6-SRC1(678-692)) IN COMPLEX WITH A TRICYCLIC INVERSE AGONIST | | Descriptor: | GLYCEROL, NUCLEAR RECEPTOR COACTIVATOR 1 CHIMERA, trans-4-[(3aR,9bR)-9b-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-1,2,3a,4,5,9b-hexahydro-3H-benzo[e]indole-3-carbonyl]cyclohexane-1-carboxylic acid | | Authors: | Sack, J. | | Deposit date: | 2019-08-19 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Rationally Designed, Conformationally Constrained Inverse Agonists of ROR gamma t-Identification of a Potent, Selective Series with Biologic-Like in Vivo Efficacy.
J.Med.Chem., 62, 2019
|
|
4UBS
 
 | | The crystal structure of cytochrome P450 105D7 from Streptomyces avermitilis in complex with Diclofenac | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Xu, L.H, Ikeda, H, Arakawa, T, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2014-08-13 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the 4'-hydroxylation of diclofenac by a microbial cytochrome P450 monooxygenase.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4OP3
 
 | | Human GKRP bound to AMG-5112 and Sorbitol-6-phosphate | | Descriptor: | (2S)-2-{6'-[(6-aminopyridin-3-yl)sulfonyl]-2'-(phenylamino)-2,3'-bipyridin-5-yl}-1,1,1-trifluoropropan-2-ol, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | Jordan, S.R, Chmait, S. | | Deposit date: | 2014-02-04 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Small Molecule Disruptors of the Glucokinase-Glucokinase Regulatory Protein Interaction: 5. A Novel Aryl Sulfone Series, Optimization Through Conformational Analysis.
J.Med.Chem., 58, 2015
|
|
4OP1
 
 | | GKRP bound to AMG0556 and Sorbitol-6-Phosphate | | Descriptor: | (2S)-2-(6-{5-[(6-aminopyridin-3-yl)sulfonyl]thiophen-2-yl}-5-chloropyridin-3-yl)-1,1,1-trifluoropropan-2-ol, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | Jordan, S.R, Chmait, S. | | Deposit date: | 2014-02-04 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Small Molecule Disruptors of the Glucokinase-Glucokinase Regulatory Protein Interaction: 5. A Novel Aryl Sulfone Series, Optimization Through Conformational Analysis.
J.Med.Chem., 58, 2015
|
|
