1NHR
 
 | |
2BC0
 
 | |
2BCP
 
 | |
4JNK
 
 | | Lactate Dehydrogenase A in complex with inhibitor compound 22 | | Descriptor: | (2R)-2-{[5-cyano-4-(3,4-dichlorophenyl)-6-oxo-1,6-dihydropyrimidin-2-yl]sulfanyl}-N-(4-sulfamoylphenyl)propanamide, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Identification of substituted 2-thio-6-oxo-1,6-dihydropyrimidines as inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2BC1
 
 | |
1TRH
 
 | |
2IRX
 
 | |
2IRU
 
 | |
2IRY
 
 | |
2H3G
 
 | |
1THG
 
 | | 1.8 ANGSTROMS REFINED STRUCTURE OF THE LIPASE FROM GEOTRICHUM CANDIDUM | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Schrag, J.D, Cygler, M. | | Deposit date: | 1992-07-28 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 A refined structure of the lipase from Geotrichum candidum.
J.Mol.Biol., 230, 1993
|
|
4JP4
 
 | | Mmp13 in complex with a reverse hydroxamate Zn-binder | | Descriptor: | CALCIUM ION, Collagenase 3, N-[(2S)-4-(5-fluoropyrimidin-2-yl)-1-({4-[5-(2,2,2-trifluoroethoxy)pyrimidin-2-yl]piperazin-1-yl}sulfonyl)butan-2-yl]-N-hydroxyformamide, ... | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2013-03-19 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Hydantoin based inhibitors of MMP13--discovery of AZD6605.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4JPA
 
 | | Mmp13 in complex with a piperazine hydantoin ligand | | Descriptor: | 3-[({2-[4-({[(4S)-4-methyl-2,5-dioxoimidazolidin-4-yl]methyl}sulfonyl)piperazin-1-yl]pyrimidin-5-yl}oxy)methyl]benzonitrile, CALCIUM ION, Collagenase 3, ... | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2013-03-19 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydantoin based inhibitors of MMP13--discovery of AZD6605.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1YQZ
 
 | | Structure of Coenzyme A-Disulfide Reductase from Staphylococcus aureus refined at 1.54 Angstrom resolution | | Descriptor: | CHLORIDE ION, COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mallett, T.C, Wallen, J.R, Sakai, H, Luba, J, Parsonage, D, Karplus, P.A, Tsukihara, T, Claiborne, A. | | Deposit date: | 2005-02-02 | | Release date: | 2006-05-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of coenzyme A-disulfide reductase from Staphylococcus aureus at 1.54 A resolution.
Biochemistry, 45, 2006
|
|
2NAX
 
 | | Structure of CCHC zinc finger domain of Pcf11 | | Descriptor: | Protein PCF11, ZINC ION | | Authors: | Yang, F, Varani, G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C terminus of Pcf11 forms a novel zinc-finger structure that plays an essential role in mRNA 3'-end processing.
RNA, 23, 2017
|
|
2NW0
 
 | | Crystal structure of a lysin | | Descriptor: | ACETATE ION, PlyB | | Authors: | Porter, C.J, Buckle, A.M, Whisstock, J.C. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A Crystal Structure of the Catalytic Domain of PlyB, a Bacteriophage Lysin Active Against Bacillus anthracis.
J.Mol.Biol., 366, 2007
|
|
2RGH
 
 | | Structure of Alpha-Glycerophosphate Oxidase from Streptococcus sp.: A Template for the Mitochondrial Alpha-Glycerophosphate Dehydrogenase | | Descriptor: | Alpha-Glycerophosphate Oxidase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Colussi, T, Boles, W, Mallett, T.C, Karplus, P.A, Claiborne, A. | | Deposit date: | 2007-10-01 | | Release date: | 2008-01-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of alpha-glycerophosphate oxidase from Streptococcus sp.: a template for the mitochondrial alpha-glycerophosphate dehydrogenase.
Biochemistry, 47, 2008
|
|
1H05
 
 | |
1FQ0
 
 | | KDPG ALDOLASE FROM ESCHERICHIA COLI | | Descriptor: | CITRIC ACID, KDPG ALDOLASE | | Authors: | Naismith, J.H. | | Deposit date: | 2000-09-01 | | Release date: | 2000-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Directed evolution of a new catalytic site in 2-keto-3-deoxy-6-phosphogluconate aldolase from Escherichia coli.
Structure, 9, 2001
|
|
1FWR
 
 | |
2LZ3
 
 | |
2LZ4
 
 | |
7K12
 
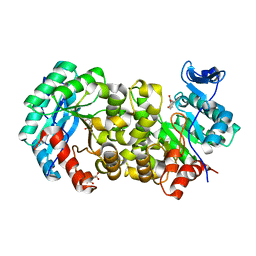 | | ACMSD in complex with diflunisal | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, 5-(2,4-DIFLUOROPHENYL)-2-HYDROXY-BENZOIC ACID, CITRIC ACID, ... | | Authors: | Yang, Y, Liu, A. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Diflunisal Derivatives as Modulators of ACMS Decarboxylase Targeting the Tryptophan-Kynurenine Pathway.
J.Med.Chem., 64, 2021
|
|
7K13
 
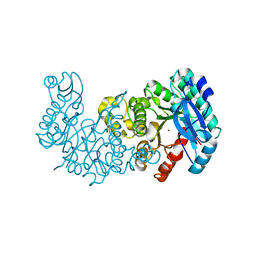 | | ACMSD in complex with diflunisal derivative 14 | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, 2-hydroxy-5-(thiophen-3-yl)benzoic acid, ZINC ION | | Authors: | Yang, Y, Liu, A. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Diflunisal Derivatives as Modulators of ACMS Decarboxylase Targeting the Tryptophan-Kynurenine Pathway.
J.Med.Chem., 64, 2021
|
|
7JIY
 
 | |
