4IXA
 
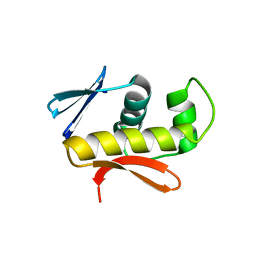 | | Structure of DNA-binding domain of the response regulator SaeR from Staphylococcus epidermidis | | Descriptor: | Response regulator SaeR | | Authors: | Chen, Y.R, Chen, S.C, Yang, C.S, Kuan, S.M, Liu, Y.H, Chen, Y. | | Deposit date: | 2013-01-24 | | Release date: | 2014-01-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of DNA-binding domain of the response regulator SaeR from Staphylococcus epidermidis
To be Published
|
|
5X07
 
 | | Crystal structure of FOXA2 DNA binding domain bound to a full consensus DNA site | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), Hepatocyte nuclear factor 3-beta | | Authors: | Li, J, Guo, M, Zhou, Z, Jiang, L, Chen, X, Qu, L, Wu, D, Chen, Z, Chen, L, Chen, Y. | | Deposit date: | 2017-01-20 | | Release date: | 2017-08-16 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structure of the Forkhead Domain of FOXA2 Bound to a Complete DNA Consensus Site
Biochemistry, 56, 2017
|
|
8EHK
 
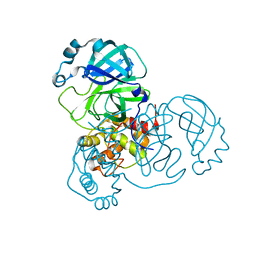 | |
8EHM
 
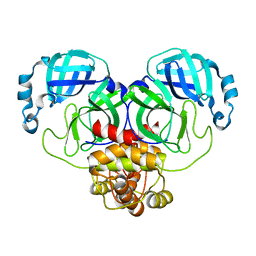 | |
8EHL
 
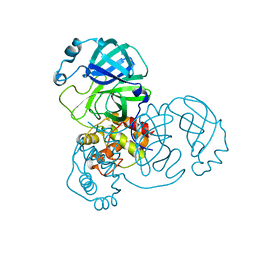 | |
8EHJ
 
 | |
1Y7H
 
 | | Structural and biochemical studies identify tobacco SABP2 as a methylsalicylate esterase and further implicate it in plant innate immunity, Northeast Structural Genomics Target AR2241 | | Descriptor: | THIOCYANATE ION, salicylic acid-binding protein 2 | | Authors: | Forouhar, F, Yang, Y, Kumar, D, Chen, Y, Fridman, E, Park, S.W, Chiang, Y, Acton, T.B, Montelione, G.T, Pichersky, E, Klessig, D.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-12-08 | | Release date: | 2004-12-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4JIS
 
 | | Crystal structure of ribitol 5-phosphate cytidylyltransferase (TarI) from Bacillus subtilis | | Descriptor: | ribitol-5-phosphate cytidylyltransferase | | Authors: | Yang, C.S, Chen, S.C, Chen, Y.R, Kuan, S.M, Liu, Y.H, Chen, Y. | | Deposit date: | 2013-03-06 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Crystal structure of ribitol 5-phosphate cytidylyltransferase (TarI) from Bacillus subtilis
To be Published
|
|
1EV7
 
 | | CRYSTAL STRUCTURE OF DNA RESTRICTION ENDONUCLEASE NAEI | | Descriptor: | TYPE IIE RESTRICTION ENDONUCLEASE NAEI | | Authors: | Huai, Q, Colandene, J.D, Chen, Y, Luo, F, Zhao, Y. | | Deposit date: | 2000-04-19 | | Release date: | 2000-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of NaeI-an evolutionary bridge between DNA endonuclease and topoisomerase.
EMBO J., 19, 2000
|
|
4BBQ
 
 | | Crystal structure of the CXXC and PHD domain of Human Lysine-specific Demethylase 2A (KDM2A)(FBXL11) | | Descriptor: | 1,2-ETHANEDIOL, LYSINE-SPECIFIC DEMETHYLASE 2A, ZINC ION | | Authors: | Allerston, C.K, Watson, A.A, Edlich, C, Li, B, Chen, Y, Ball, L, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Laue, E.D, Gileadi, O. | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of the Cxxc and Phd Domain of Human Lysine-Specific Demethylase 2A (Kdm2A)(Fbxl11)
To be Published
|
|
1FGL
 
 | | Cyclophilin A complexed with a fragment of HIV-1 GAG protein | | Descriptor: | CYCLOPHILIN A, HIV-1 GAG PROTEIN | | Authors: | Zhao, Y, Chen, Y, Schutkowski, M, Fischer, G, Ke, H. | | Deposit date: | 1996-11-18 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclophilin A complexed with a fragment of HIV-1 gag protein: insights into HIV-1 infectious activity.
Structure, 5, 1997
|
|
9J4L
 
 | | Crystal structure of GH9l Inulin fructotransferases (IFTase) | | Descriptor: | DFA-III-forming inulin fructotransferase | | Authors: | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | Deposit date: | 2024-08-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|
7T5S
 
 | | P. aeruginosa LpxA in complex with ligand H16 | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, N~2~-(cyclohexylacetyl)-N-1H-tetrazol-5-yl-L-alaninamide | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Based Ligand Design Targeting Pseudomonas aeruginosa LpxA in Lipid A Biosynthesis.
Acs Infect Dis., 8, 2022
|
|
7T5Z
 
 | | P. aeruginosa LpxA in complex with ligand L8 | | Descriptor: | (4S)-N-(1H-tetrazol-5-yl)-2-[3-(trifluoromethyl)benzene-1-sulfonyl]-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DI(HYDROXYETHYL)ETHER | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Ligand Design Targeting Pseudomonas aeruginosa LpxA in Lipid A Biosynthesis.
Acs Infect Dis., 8, 2022
|
|
7T5R
 
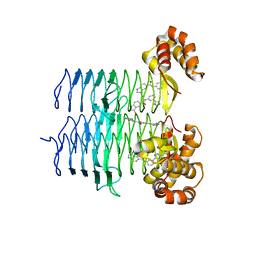 | | P. aeruginosa LpxA in complex with ligand H7 | | Descriptor: | 3-bromo-N-[3-(1H-tetrazol-5-yl)phenyl]-1H-indole-5-carboxamide, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, GLYCEROL | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Ligand Design Targeting Pseudomonas aeruginosa LpxA in Lipid A Biosynthesis.
Acs Infect Dis., 8, 2022
|
|
7T5X
 
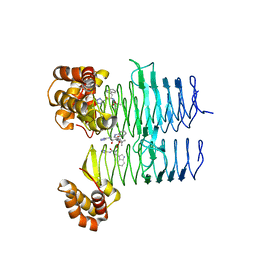 | | P. aeruginosa LpxA in complex with ligand L6 | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, Nalpha-(tert-butoxycarbonyl)-N-1H-tetrazol-5-yl-D-tryptophanamide | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Ligand Design Targeting Pseudomonas aeruginosa LpxA in Lipid A Biosynthesis.
Acs Infect Dis., 8, 2022
|
|
7T60
 
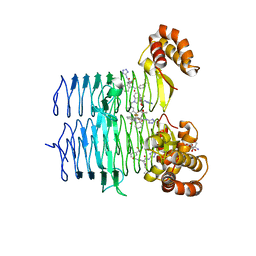 | | P. aeruginosa LpxA in complex with ligand L13 | | Descriptor: | (3S)-3-(5,5-dimethyl-2-oxo-1,3-oxazolidin-3-yl)-N-(1H-tetrazol-5-yl)-1-[3-(trifluoromethyl)benzoyl]-2,3-dihydro-1H-indole-3-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Ligand Design Targeting Pseudomonas aeruginosa LpxA in Lipid A Biosynthesis.
Acs Infect Dis., 8, 2022
|
|
7T61
 
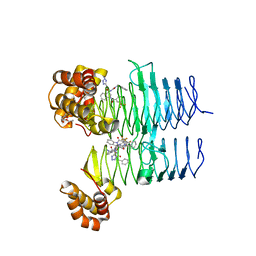 | | P. aeruginosa LpxA in complex with ligand L15 | | Descriptor: | (3S)-3-({[(Z)-phenylmethylidene]carbamoyl}amino)-N-(1H-tetrazol-5-yl)-1-[3-(trifluoromethyl)benzoyl]-2,3-dihydro-1H-indole-3-carboxamide, ACETATE ION, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Ligand Design Targeting Pseudomonas aeruginosa LpxA in Lipid A Biosynthesis.
Acs Infect Dis., 8, 2022
|
|
9J4I
 
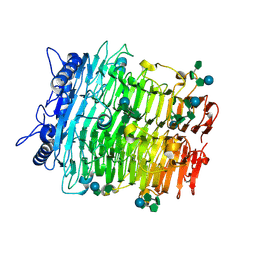 | | Crystal structure of GH9l Inulin fructotransferases (IFTase) in compex with fruetosyl nystose (GF4) | | Descriptor: | DFA-III-forming inulin fructotransferase, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-[alpha-D-glucopyranose-(1-2)]beta-D-fructofuranose, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-[alpha-D-glucopyranose-(1-2)]beta-D-fructofuranose | | Authors: | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | Deposit date: | 2024-08-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|
9J4K
 
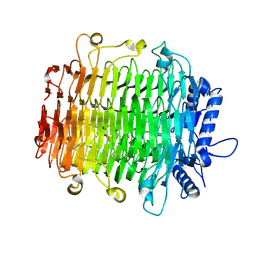 | | Crystal structure of GH9l Inulinfructotransferases (IFTase) in complex with GF2 | | Descriptor: | DFA-III-forming inulin fructotransferase, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | Deposit date: | 2024-08-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|
9J4J
 
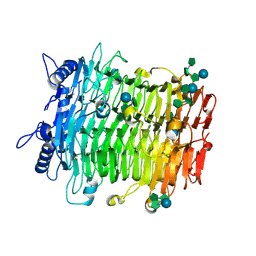 | | Crystal structure of GH9l Inulin fructotransferases(IFTase)incomplex with nystose(F3) | | Descriptor: | DFA-III-forming inulin fructotransferase, beta-D-fructofuranose, beta-D-fructofuranose-(1-1)-beta-D-fructofuranose, ... | | Authors: | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | Deposit date: | 2024-08-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|
7STA
 
 | |
1ZKL
 
 | | Multiple Determinants for Inhibitor Selectivity of Cyclic Nucleotide Phosphodiesterases | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A, MAGNESIUM ION, ... | | Authors: | Wang, H, Liu, Y, Chen, Y, Robinson, H, Ke, H. | | Deposit date: | 2005-05-03 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Multiple elements jointly determine inhibitor selectivity of cyclic nucleotide phosphodiesterases 4 and 7
J.Biol.Chem., 280, 2005
|
|
6PK0
 
 | | Crystal Structure of OXA-48 with Hydrolyzed Imipenem | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2019-06-28 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Substrate Specificity and Carbapenemase Activity of OXA-48 Class D beta-Lactamase.
Acs Infect Dis., 6, 2020
|
|
1MN9
 
 | | NDP kinase mutant (H122G) complex with RTP | | Descriptor: | MAGNESIUM ION, NDP kinase, RIBAVIRIN TRIPHOSPHATE | | Authors: | Gallois-montbrun, S, Chen, Y, Dutartre, H, Morera, S, Guerreiro, C, Mulard, L, Schneider, B, Janin, J, Canard, B, Veron, M, Deville-bonne, D. | | Deposit date: | 2002-09-05 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Analysis of the Activation of Ribavirin Analogs by NDP Kinase: Comparison with Other Ribavirin Targets
MOL.PHARMACOL., 63, 2003
|
|
