4HT8
 
 | | Crystal structure of E coli Hfq bound to poly(A) A7 | | Descriptor: | Protein hfq, RNA (5'-R(*AP*AP*AP*AP*AP*AP*A)-3') | | Authors: | Wang, W.W, Wang, L.J. | | Deposit date: | 2012-11-01 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hfq-bridged ternary complex is important for translation activation of rpoS by DsrA.
Nucleic Acids Res., 41, 2013
|
|
4HT9
 
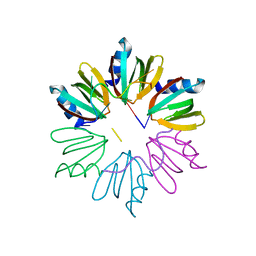 | | Crystal structure of E coli Hfq bound to two RNAs | | Descriptor: | Protein hfq, RNA (5'-R(*AP*AP*AP*AP*AP*AP*A)-3'), RNA (5'-R(*AP*UP*UP*UP*UP*UP*UP*A)-3') | | Authors: | Wang, W.W, Wang, L.J. | | Deposit date: | 2012-11-01 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hfq-bridged ternary complex is important for translation activation of rpoS by DsrA.
Nucleic Acids Res., 41, 2013
|
|
3Q0T
 
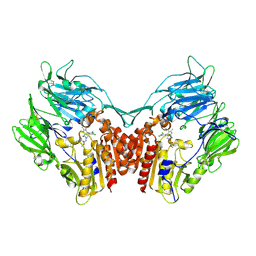 | |
5ZND
 
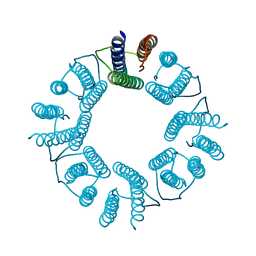 | | 8-mer nanotube derived from 24-mer rHuHF nanocage | | Descriptor: | Ferritin heavy chain | | Authors: | Wang, W.M, Wang, L.L, Zang, J.C, Chen, H, Zhao, G.H, Wang, H.F. | | Deposit date: | 2018-04-09 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Selective Elimination of the Key Subunit Interfaces Facilitates Conversion of Native 24-mer Protein Nanocage into 8-mer Nanorings.
J. Am. Chem. Soc., 140, 2018
|
|
3SWW
 
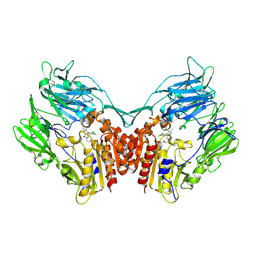 | | Crystal structure of human dpp-iv in complex with sa-(+)-3-(aminomethyl)-4-(2,4-dichlorophenyl)-6-(2-methoxyphenyl)- 2-methyl-5h-pyrrolo[3,4-b]pyridin-7(6h)-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(aminomethyl)-4-(2,4-dichlorophenyl)-6-(2-methoxyethyl)-2-methyl-5,6-dihydro-7H-pyrrolo[3,4-b]pyridin-7-one, ... | | Authors: | Klei, H.E. | | Deposit date: | 2011-07-14 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 7-Oxopyrrolopyridine-derived DPP4 inhibitors-mitigation of CYP and hERG liabilities via introduction of polar functionalities in the active site.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SX4
 
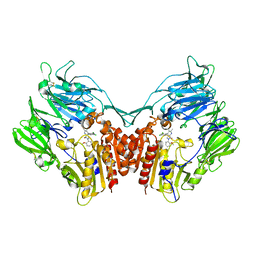 | | Crystal structure of human dpp-iv in complex with sa-(+)-3-(aminomethyl)-4-(2,4-dichlorophenyl)-6-(2-methoxyphenyl)- 2-methyl-5h-pyrrolo[3,4-b]pyridin-7(6h)-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(aminomethyl)-4-(2,4-dichlorophenyl)-6-(2-methoxyphenyl)-2-methyl-5,6-dihydro-7H-pyrrolo[3,4-b]pyridin-7-one, ... | | Authors: | Klei, H.E. | | Deposit date: | 2011-07-14 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 7-Oxopyrrolopyridine-derived DPP4 inhibitors-mitigation of CYP and hERG liabilities via introduction of polar functionalities in the active site.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7FAP
 
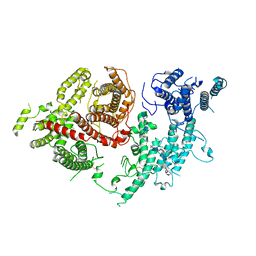 | | Structure of VAR2CSA-CSA 3D7 | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-3)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, Erythrocyte membrane protein 1, PfEMP1 | | Authors: | Wang, L, Wang, Z. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The molecular mechanism of cytoadherence to placental or tumor cells through VAR2CSA from Plasmodium falciparum.
Cell Discov, 7, 2021
|
|
7WRG
 
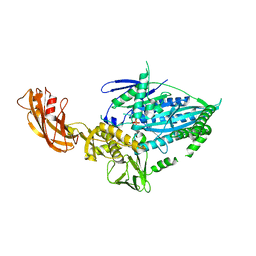 | | Crystal structure of full-length kinesin-3 KLP-6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein, MAGNESIUM ION | | Authors: | Wang, W.J, Ren, J.Q, Song, W.Y, Feng, W. | | Deposit date: | 2022-01-26 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | The architecture of kinesin-3 KLP-6 reveals a multilevel-lockdown mechanism for autoinhibition.
Nat Commun, 13, 2022
|
|
5HAD
 
 | |
8JMX
 
 | |
7FAS
 
 | | VAR2CSA 3D7 ectodomain core region | | Descriptor: | Erythrocyte membrane protein 1, PfEMP1 | | Authors: | Wang, L, Zhaoning, W. | | Deposit date: | 2021-07-07 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The molecular mechanism of cytoadherence to placental or tumor cells through VAR2CSA from Plasmodium falciparum.
Cell Discov, 7, 2021
|
|
5VA3
 
 | |
5VA2
 
 | |
5VA1
 
 | |
3CTB
 
 | | Tethered PXR-LBD/SRC-1p apoprotein | | Descriptor: | Pregnane X receptor, Linker, Steroid receptor coactivator 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2008-04-11 | | Release date: | 2008-12-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Construction and characterization of a fully active PXR/SRC-1 tethered protein with increased stability
Protein Eng.Des.Sel., 21, 2008
|
|
2REW
 
 | | Crystal Structure of PPARalpha ligand binding domain with BMS-631707 | | Descriptor: | (2S,3S)-1-(4-METHOXYPHENYL)-3-(3-(2-(5-METHYL-2-PHENYLOXAZOL-4-YL)ETHOXY)BENZYL)-4-OXOAZETIDINE-2-CARBOXYLIC ACID, N,N-BIS(3-D-GLUCONAMIDOPROPYL)DEOXYCHOLAMIDE, Peroxisome proliferator-activated receptor alpha | | Authors: | Muckelbauer, J. | | Deposit date: | 2007-09-27 | | Release date: | 2007-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Azetidinone Acids as Conformationally-Constrained Dual (alpha/gamma) PPAR Activators
To be Published
|
|
6IVE
 
 | |
6KE4
 
 | | ABloop reengineered Ferritin Nanocage | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Wang, W.M, Wang, H.F. | | Deposit date: | 2019-07-03 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AB loop engineered ferritin nanocages for drug loading under benign experimental conditions.
Chem.Commun.(Camb.), 55, 2019
|
|
6KE2
 
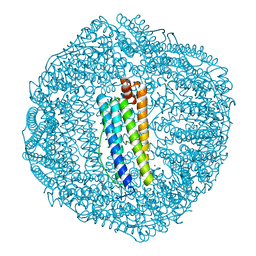 | | ABloop reengineered Ferritin Nanocage | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Wang, W.M, Wang, H.F. | | Deposit date: | 2019-07-03 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | AB loop engineered ferritin nanocages for drug loading under benign experimental conditions.
Chem.Commun.(Camb.), 55, 2019
|
|
4EFG
 
 | |
4EXZ
 
 | |
4EDE
 
 | |
4EEJ
 
 | |
7DIE
 
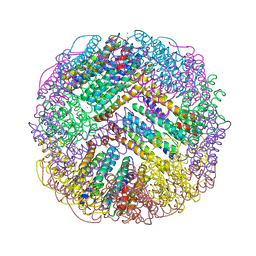 | | Crystal structure of M. penetrans Ferritin | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Wang, w.m, Zhang, y, Wang, h.f. | | Deposit date: | 2020-11-19 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ferritin with Atypical Ferroxidase Centers Takes B-Channels as the Pathway for Fe 2+ Uptake from Mycoplasma .
Inorg.Chem., 60, 2021
|
|
7DWO
 
 | |
