3JXS
 
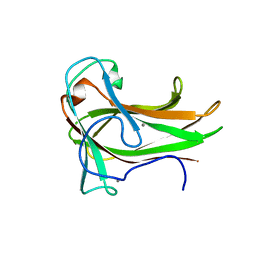 | | Crystal structure of XG34, an evolved xyloglucan binding CBM | | Descriptor: | ACETATE ION, CALCIUM ION, Xylanase | | Authors: | Divne, C, Tan, T.-C, Brumer, H, Gullfot, F. | | Deposit date: | 2009-09-21 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of XG-34, an evolved xyloglucan-specific carbohydrate-binding module.
Proteins, 78, 2009
|
|
3J8Y
 
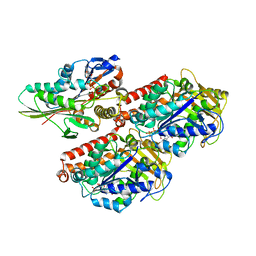 | | High-resolution structure of ATP analog-bound kinesin on microtubules | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shang, Z, Zhou, K, Xu, C, Csencsits, R, Cochran, J.C, Sindelar, C.V. | | Deposit date: | 2014-11-20 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation.
Elife, 3, 2014
|
|
3K1N
 
 | |
3K1K
 
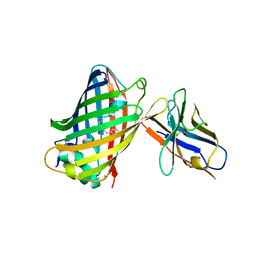 | | Green fluorescent protein bound to enhancer nanobody | | Descriptor: | Enhancer, Green Fluorescent Protein | | Authors: | Kirchhofer, A, Helma, J, Schmidthals, K, Frauer, C, Cui, S, Karcher, A, Pellis, M, Muyldermans, S, Delucci, C.C, Cardoso, M.C, Leonhardt, H, Hopfner, K.-P, Rothbauer, U. | | Deposit date: | 2009-09-28 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Modulation of protein properties in living cells using nanobodies
Nat.Struct.Mol.Biol., 17, 2010
|
|
6EY5
 
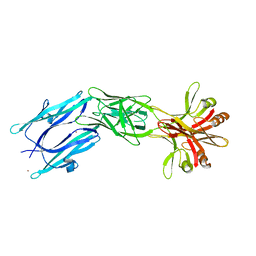 | | C-terminal part (residues 224-515) of PorM | | Descriptor: | T9SS component cytoplasmic membrane protein PorM, ZINC ION | | Authors: | Leone, P, Cambillau, C, Roussel, A. | | Deposit date: | 2017-11-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Type IX secretion system PorM and gliding machinery GldM form arches spanning the periplasmic space.
Nat Commun, 9, 2018
|
|
6EWS
 
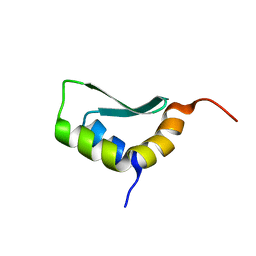 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12A-NDD | | Descriptor: | NRPS Kj12A-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
1TZB
 
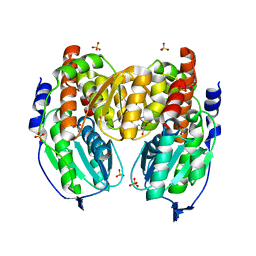 | | Crystal structure of native phosphoglucose/phosphomannose isomerase from Pyrobaculum aerophilum | | Descriptor: | GLYCEROL, SULFATE ION, glucose-6-phosphate isomerase, ... | | Authors: | Swan, M.K, Hansen, T, Schoenheit, P, Davies, C. | | Deposit date: | 2004-07-09 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | A novel phosphoglucose/phosphomannose isomease from the crenarchaeon Pyrobaculum aerophilum is a member of the PGI superfamily: structural evidence at 1.16 A resolution
J.Biol.Chem., 279, 2004
|
|
6F7L
 
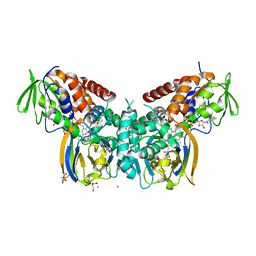 | | Crystal structure of LkcE R326Q mutant in complex with its substrate | | Descriptor: | ACETATE ION, Amine oxidase LkcE, CALCIUM ION, ... | | Authors: | Dorival, J, Risser, F, Jacob, C, Collin, S, Drager, G, Kirschning, A, Paris, C, Chagot, B, Gruez, A, Weissman, K.J. | | Deposit date: | 2017-12-11 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into a dual function amide oxidase/macrocyclase from lankacidin biosynthesis.
Nat Commun, 9, 2018
|
|
1UFL
 
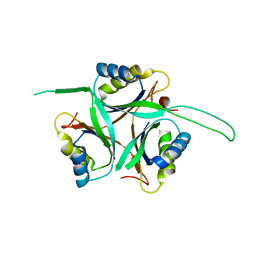 | | Crystal Structure of TT1020 from Thermus thermophilus HB8 | | Descriptor: | Nitrogen regulatory protein P-II | | Authors: | Wang, H, Sakai, H, Hori-Takemoto, C, Kaminishi, T, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-31 | | Release date: | 2003-11-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the signal transducing protein GlnK from Thermus thermophilus HB8.
J.Struct.Biol., 149, 2005
|
|
6FBA
 
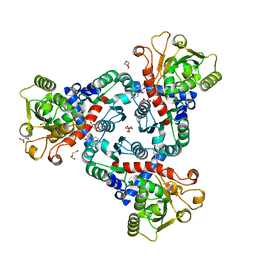 | | Crystal Structure of truncated aspartate transcarbamoylase from Plasmodium falciparum with bound inhibitor 2,3-naphthalenediol | | Descriptor: | Aspartate transcarbamoylase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Lunev, S, Bosch, S.S, Batista, F.A, Wang, C, Wrenger, C, Groves, M.R. | | Deposit date: | 2017-12-18 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a non-competitive inhibitor of Plasmodium falciparum aspartate transcarbamoylase.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
6EWV
 
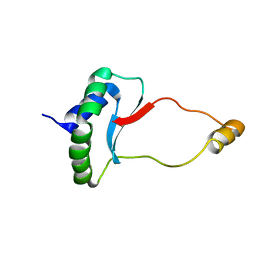 | | Solution Structure of Docking Domain Complex of RXP NRPS: Kj12C NDD - Kj12B CDD | | Descriptor: | NRPS Kj12C-NDD, NRPS Kj12B-CDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6FIC
 
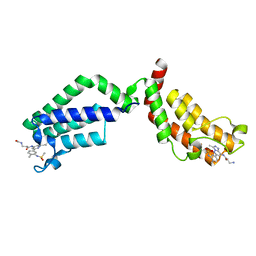 | | Bivalent Inhibitor UNC4512 Bound to the TAF1 Bromodomain Tandem | | Descriptor: | 3-azanyl-2-[3-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[[3-azanyl-1-[[2-[[3-methyl-6-[4-methyl-3-(methylsulfonyl-$l^{2}-azanyl)cyclohexa-1,3,5-trien-1-yl]-[1,2,4]triazolo[4,3-b]pyridazin-8-yl]-$l^{2}-azanyl]-2-oxidanylidene-ethyl]amino]-1-oxidanylidene-propan-2-yl]amino]-2-oxidanylidene-ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]propanoylamino]-~{N}-[3-[[3-methyl-6-[4-methyl-3-(methylsulfonylamino)phenyl]-[1,2,4]triazolo[4,3-b]pyridazin-8-yl]amino]-3-oxidanylidene-propyl]propanamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Mathea, S, Suh, J.L, Salah, E, Tallant, C, Siejka, P, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, James, L.I, Frye, S.V, Knapp, S. | | Deposit date: | 2018-01-17 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Bivalent Inhibitor UNC4512 Bound to the TAF1 Bromodomain Tandem
To Be Published
|
|
6KXR
 
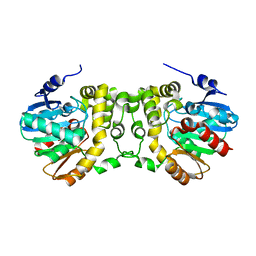 | | Crystal structure of wild type Alp1U from the biosynthesis of kinamycins | | Descriptor: | D-MALATE, Putative hydrolase | | Authors: | Zhang, L, Yingli, Z, De, B.C, Zhang, C. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.45073676 Å) | | Cite: | Mutation of an atypical oxirane oxyanion hole improves regioselectivity of the alpha / beta-fold epoxide hydrolase Alp1U.
J.Biol.Chem., 295, 2020
|
|
6KY8
 
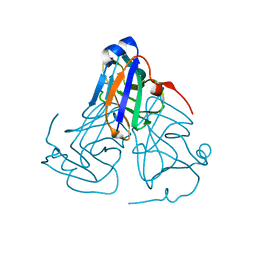 | | Crystal structure of ASFV dUTPase | | Descriptor: | E165R | | Authors: | Li, C, Chai, Y, Song, H, Qi, J, Sun, Y, Gao, G.F. | | Deposit date: | 2019-09-17 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal Structure of African Swine Fever Virus dUTPase Reveals a Potential Drug Target.
Mbio, 10, 2019
|
|
7N4K
 
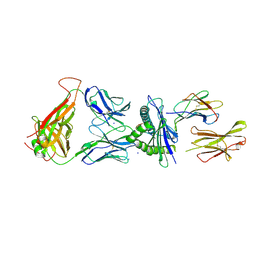 | | 6218 TCR in complex with H2-Db PA 224 | | Descriptor: | Beta-2-microglobulin, Fusion protein of T cell receptor alpha variable 21-DV12 and T-cell receptor, sp3.4 alpha chain, ... | | Authors: | Szeto, C, Gras, S. | | Deposit date: | 2021-06-04 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Covalent TCR-peptide-MHC interactions induce T cell activation and redirect T cell fate in the thymus.
Nat Commun, 13, 2022
|
|
7N5C
 
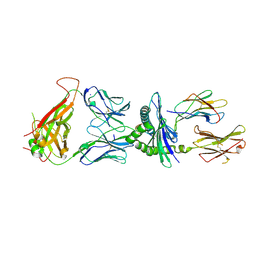 | | 6218 TCR in complex with H2Db PA with an engineered TCR-pMHC disulfide bond | | Descriptor: | Beta-2-microglobulin, Fusion protein of T cell receptor alpha variable 21-DV12 with T-cell receptor, sp3.4 alpha chain, ... | | Authors: | Szeto, C, Gras, S. | | Deposit date: | 2021-06-05 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Covalent TCR-peptide-MHC interactions induce T cell activation and redirect T cell fate in the thymus.
Nat Commun, 13, 2022
|
|
7N5P
 
 | | 6218 TCR in complex with H2-Db PA224-233 with a cysteine mutant | | Descriptor: | Beta-2-microglobulin, Fusion protein of T cell receptor alpha variable 21-DV12 and T-cell receptor, sp3.4 alpha chain, ... | | Authors: | Szeto, C, Gras, S. | | Deposit date: | 2021-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Covalent TCR-peptide-MHC interactions induce T cell activation and redirect T cell fate in the thymus.
Nat Commun, 13, 2022
|
|
7N5Q
 
 | |
7N8D
 
 | |
7N57
 
 | |
7N4O
 
 | |
7N8G
 
 | | Crystal structure of R20A-R22A human Galectin-7 mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2021-06-14 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of R20A human Galectin-7 mutant
To Be Published
|
|
7N6C
 
 | | Crystal structure of R22A human Galectin-7 mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2021-06-08 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of R20A human Galectin-7 mutant
To Be Published
|
|
7N96
 
 | |
7NMQ
 
 | | Bacillus cereus HblL1 toxin component | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Hemolysin BL lytic component L1 | | Authors: | Rizkallah, P.J, Berry, C. | | Deposit date: | 2021-02-23 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The Crystal Structure of Bacillus cereus HblL 1 .
Toxins, 13, 2021
|
|
