1K99
 
 | |
1CXR
 
 | |
5B7N
 
 | |
5B7P
 
 | |
5B7G
 
 | |
5B7Q
 
 | |
3DW8
 
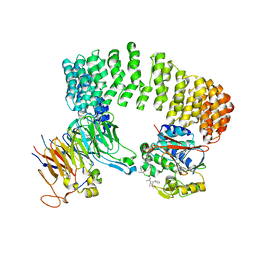 | | Structure of a Protein Phosphatase 2A Holoenzyme with B55 subunit | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Xu, Y, Chen, Y, Zhang, P, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2008-07-21 | | Release date: | 2008-10-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of a protein phosphatase 2A holoenzyme: insights into B55-mediated Tau dephosphorylation.
Mol.Cell, 31, 2008
|
|
2NPP
 
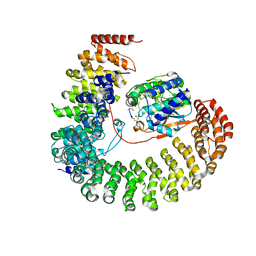 | | Structure of the Protein Phosphatase 2A Holoenzyme | | Descriptor: | MANGANESE (II) ION, Protein Phosphatase 2, regulatory subunit A (PR 65), ... | | Authors: | Xu, Y, Chen, Y, Xing, Y, Chao, Y, Shi, Y. | | Deposit date: | 2006-10-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the protein phosphatase 2A holoenzyme
Cell(Cambridge,Mass.), 127, 2006
|
|
1WP7
 
 | | crystal structure of Nipah Virus fusion core | | Descriptor: | fusion protein | | Authors: | Xu, Y, Liu, Y, Lou, Z, Su, N, Bai, Z, Gao, G.F, Rao, Z. | | Deposit date: | 2004-08-31 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Nipah Virus fusion core
To be Published
|
|
1WNC
 
 | | Crystal structure of the SARS-CoV Spike protein fusion core | | Descriptor: | E2 glycoprotein | | Authors: | Xu, Y, Lou, Z, Liu, Y, Pang, H, Tien, P, Gao, G.F, Rao, Z. | | Deposit date: | 2004-07-29 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core
J.Biol.Chem., 279, 2004
|
|
2EEP
 
 | | Prolyl Tripeptidyl Aminopeptidase Complexed with an Inhibitor | | Descriptor: | Dipeptidyl aminopeptidase IV, putative, SULFATE ION, ... | | Authors: | Xu, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2007-02-16 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel inhibitor for prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis and details of substrate-recognition mechanism
J.Mol.Biol., 375, 2008
|
|
4J0E
 
 | |
4J0F
 
 | |
4DK1
 
 | |
4DK0
 
 | |
6VWI
 
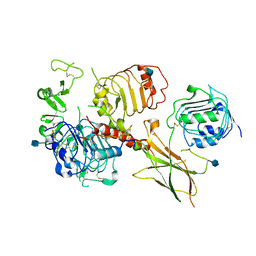 | | Head region of the closed conformation of the human type 1 insulin-like growth factor receptor ectodomain in complex with human insulin-like growth factor II. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin-like growth factor II, Leucine-zippered human type 1 insulin-like growth factor receptor ectodomain | | Authors: | Xu, Y, Kirk, N.S, Lawrence, M.C, Croll, T.I. | | Deposit date: | 2020-02-19 | | Release date: | 2020-05-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | How IGF-II Binds to the Human Type 1 Insulin-like Growth Factor Receptor.
Structure, 28, 2020
|
|
6VWG
 
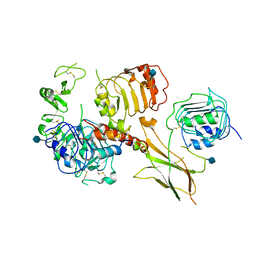 | | Head region of the open conformation of the human type 1 insulin-like growth factor receptor ectodomain in complex with human insulin-like growth factor II. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin-like growth factor II, Leucine-zippered human type 1 insulin-like growth factor receptor ectodomain | | Authors: | Xu, Y, Kirk, N.S, Lawrence, M.C, Croll, T.I. | | Deposit date: | 2020-02-19 | | Release date: | 2020-05-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | How IGF-II Binds to the Human Type 1 Insulin-like Growth Factor Receptor.
Structure, 28, 2020
|
|
6VWJ
 
 | |
1NH5
 
 | | AUTOMATIC ASSIGNMENT OF NMR DATA AND DETERMINATION OF THE PROTEIN STRUCTURE OF A NEW WORLD SCORPION NEUROTOXIN USING NOAH/DIAMOD | | Descriptor: | Neurotoxin 5 | | Authors: | Xu, Y, Jablonsky, M.J, Jackson, P.L, Krishna, N.R, Braun, W. | | Deposit date: | 2002-12-18 | | Release date: | 2003-01-07 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | Automatic assignment of NOESY Cross peaks and determination of the protein structure of a new world scorpion neurotoxin Using NOAH/DIAMOD
J.Magn.Reson., 148, 2001
|
|
6VWH
 
 | |
1QR6
 
 | | HUMAN MITOCHONDRIAL NAD(P)-DEPENDENT MALIC ENZYME | | Descriptor: | MALIC ENZYME 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Xu, Y, Bhargava, G, Wu, H, Loeber, G, Tong, L. | | Deposit date: | 1999-06-18 | | Release date: | 1999-07-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human mitochondrial NAD(P)(+)-dependent malic enzyme: a new class of oxidative decarboxylases.
Structure, 7, 1999
|
|
2Z3W
 
 | | Prolyl tripeptidyl aminopeptidase mutant E636A | | Descriptor: | Dipeptidyl aminopeptidase IV, GLYCEROL, SULFATE ION | | Authors: | Xu, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2007-06-07 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel inhibitor for prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis and details of substrate-recognition mechanism
J.Mol.Biol., 375, 2008
|
|
2Z3Z
 
 | | Prolyl tripeptidyl aminopeptidase mutant E636A complexd with an inhibitor | | Descriptor: | Dipeptidyl aminopeptidase IV, SULFATE ION, [(2R)-1-(L-ALANYL-L-ISOLEUCYL)PYRROLIDIN-2-YL]BORONIC ACID | | Authors: | Xu, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2007-06-09 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel inhibitor for prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis and details of substrate-recognition mechanism
J.Mol.Biol., 375, 2008
|
|
1QY7
 
 | | The structure of the PII protein from the cyanobacteria Synechococcus sp. PCC 7942 | | Descriptor: | NICKEL (II) ION, Nitrogen regulatory protein P-II, SULFATE ION | | Authors: | Xu, Y, Carr, P.D, Clancy, P, Garcia-Dominguez, M, Forchhammer, K, Florencio, F, Tandeau de Marsac, N, Vasudevan, S.G, Ollis, D.L. | | Deposit date: | 2003-09-09 | | Release date: | 2003-09-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of the PII proteins from the cyanobacteria Synechococcus sp. PCC 7942 and Synechocystis sp. PCC 6803.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2H25
 
 | |
