1UQS
 
 | | The Crystal Structure of Human CD1b with a Bound Bacterial Glycolipid | | Descriptor: | BETA-2-MICROGLOBULIN, GLUCOSE MONOMYCOLATE, T-CELL SURFACE GLYCOPROTEIN CD1B | | Authors: | Batuwangala, T, Shepherd, D, Gadola, S.D, Gibson, K.J.C, Zaccai, N.R, Besra, G.S, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2003-10-16 | | Release date: | 2003-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The crystal structure of human CD1b with a bound bacterial glycolipid.
J Immunol., 172, 2004
|
|
2L7Y
 
 | |
2LI6
 
 | |
7DYG
 
 | |
7DYQ
 
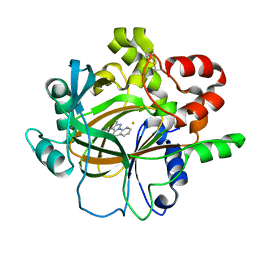 | | Crystal structure of histone lysine demethylase 4D (KDM4D) in complex with the inhibitor 5-hydroxy-2-methylpyrazolo[1,5-a]pyrido[3,2-e]pyrimidine-3-carbonitrile | | Descriptor: | 5-hydroxy-2-methylpyrazolo[1,5-a]pyrido[3,2-e]pyrimidine-3-carbonitrile, FE (III) ION, Lysine-specific demethylase 4D | | Authors: | Wang, T, Yang, L. | | Deposit date: | 2021-01-22 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of histone lysine demethylase 4D (KDM4D) in complex with the inhibitor 5-hydroxy-2-methylpyrazolo[1,5-a]pyrido[3,2-e]pyrimidine-3-carbonitrile
To Be Published
|
|
2H3B
 
 | | Crystal Structure of Mouse Nicotinamide Phosphoribosyltransferase/Visfatin/Pre-B Cell Colony Enhancing Factor 1 | | Descriptor: | Nicotinamide phosphoribosyltransferase, SULFATE ION | | Authors: | Wang, T, Zhang, X, Bheda, P, Revollo, J.R, Imai, S.I, Wolberger, C. | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-20 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Nampt/PBEF/visfatin, a mammalian NAD(+) biosynthetic enzyme.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2H3D
 
 | | Crystal Structure of Mouse Nicotinamide Phosphoribosyltransferase/Visfatin/Pre-B Cell Colony Enhancing Factor in Complex with Nicotinamide Mononuleotide | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide phosphoribosyltransferase | | Authors: | Wang, T, Zhang, X, Bheda, P, Revollo, J.R, Imai, S.I, Wolberger, C. | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Nampt/PBEF/visfatin, a mammalian NAD(+) biosynthetic enzyme.
Nat.Struct.Mol.Biol., 13, 2006
|
|
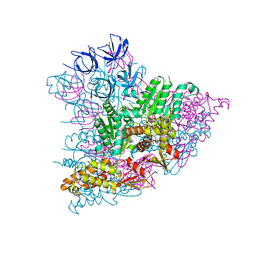
5KWA
 
 | |
6WKS
 
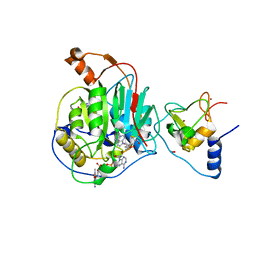 | | Structure of SARS-CoV-2 nsp16/nsp10 in complex with RNA cap analogue (m7GpppA) and S-adenosylmethionine | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase, ADENOSINE, ... | | Authors: | Gupta, Y.K, Viswanathan, T, Arya, S, Qi, S, Misra, A, Chan, S.-H. | | Deposit date: | 2020-04-16 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of RNA cap modification by SARS-CoV-2.
Nat Commun, 11, 2020
|
|
4LUQ
 
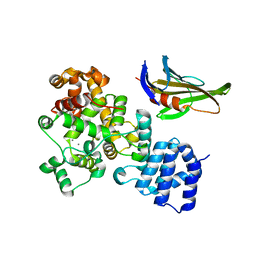 | |
6ES9
 
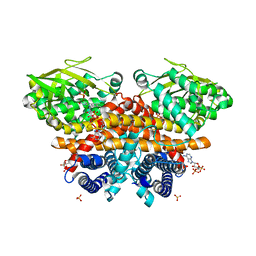 | | Methylsuccinyl-CoA dehydrogenase of Paracoccus denitrificans with bound flavin adenine dinucleotide | | Descriptor: | Acyl-CoA dehydrogenase, COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zarzycki, J, Schwander, T, Erb, T.J. | | Deposit date: | 2017-10-19 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural basis for substrate specificity of methylsuccinyl-CoA dehydrogenase, an unusual member of the acyl-CoA dehydrogenase family.
J. Biol. Chem., 293, 2018
|
|
7LW3
 
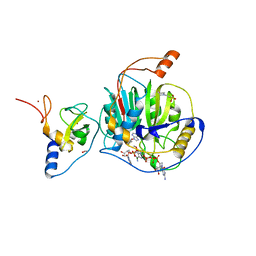 | | Structure of SARS-CoV-2 nsp16/nsp10 complex in presence of Cap-1 analog (m7GpppAmU) and SAH | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Gupta, Y.K, Viswanathan, T, Misra, A, Qi, S. | | Deposit date: | 2021-02-27 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A metal ion orients SARS-CoV-2 mRNA to ensure accurate 2'-O methylation of its first nucleotide.
Nat Commun, 12, 2021
|
|
7LW4
 
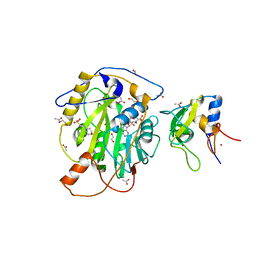 | | Structure of SARS-CoV-2 nsp16/nsp10 complex in presence of S-adenosyl-L-homocysteine (SAH) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Gupta, Y.K, Viswanathan, T, Misra, A, Qi, S. | | Deposit date: | 2021-02-27 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A metal ion orients SARS-CoV-2 mRNA to ensure accurate 2'-O methylation of its first nucleotide.
Nat Commun, 12, 2021
|
|
3KCK
 
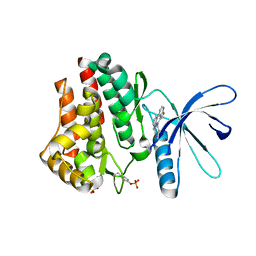 | | A Novel Chemotype of Kinase Inhibitors | | Descriptor: | 3-chloro-4-(4H-3,4,7-triazadibenzo[cd,f]azulen-6-yl)phenol, Tyrosine-protein kinase JAK2 | | Authors: | Zuccola, H.J, Wang, T, Ledeboer, M.W. | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A novel chemotype of kinase inhibitors: Discovery of 3,4-ring fused 7-azaindoles and deazapurines as potent JAK2 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3M9H
 
 | |
3M9B
 
 | |
3M9D
 
 | |
3FP9
 
 | |
3OHN
 
 | | Crystal structure of the FimD translocation domain | | Descriptor: | Outer membrane usher protein FimD | | Authors: | Wang, T, Li, H. | | Deposit date: | 2010-08-17 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate.
Nature, 474, 2011
|
|
4EFB
 
 | | Crystal structure of DNA ligase | | Descriptor: | 4-amino-2-(cyclopentyloxy)-6-{[(1R,2S)-2-hydroxycyclopentyl]oxy}pyrimidine-5-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Wei, Y, Wang, T, Charifson, P, Xu, W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of DNA ligase
To be Published
|
|
4DOX
 
 | |
4FQB
 
 | |
4EFE
 
 | | crystal structure of DNA ligase | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, SULFATE ION, ... | | Authors: | Wei, Y, Wang, T, Charifson, P, Xu, W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of DNA ligase
To be Published
|
|
4O56
 
 | | Structure of PLK1 in complex with peptide | | Descriptor: | PHOSPHATE ION, Serine/threonine-protein kinase PLK1, synthetic peptide | | Authors: | Wang, T. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Integration of Charge-dipole Interaction and Intramolecular Hydrogen Bond in Ligand Design for the Polo-Box Domain of Polo-like Kinase 1
To be Published
|
|
3WA5
 
 | | Crystal Structure of type VI peptidoglycan muramidase effector Tse3 in complex with its cognate immunity protein Tsi3 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Tse3-specific immunity protein, ... | | Authors: | Ding, J, Wang, T, Liu, W, Wang, D.C. | | Deposit date: | 2013-04-26 | | Release date: | 2013-10-02 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Complex structure of type VI peptidoglycan muramidase effector and a cognate immunity protein.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
