2QD8
 
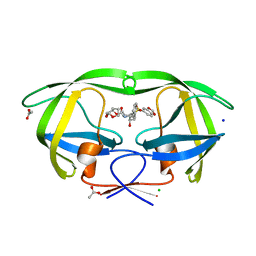 | | HIV-1 Protease Mutant I84V with potent Antiviral inhibitor GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.F, Tie, Y, Boross, P.I, Tozser, J, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2007-06-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.
J.Med.Chem., 50, 2007
|
|
2QD7
 
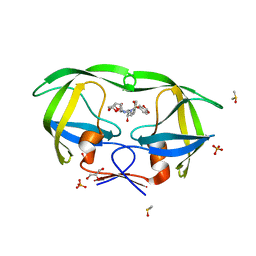 | | HIV-1 Protease Mutant V82A with potent Antiviral inhibitor GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wang, Y.F, Tie, Y, Boross, P.I, Tozser, J, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2007-06-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.
J.Med.Chem., 50, 2007
|
|
2QD6
 
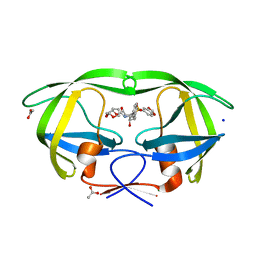 | | HIV-1 Protease Mutant I50V with potent Antiviral inhibitor GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.F, Tie, Y, Boross, P.I, Tozser, J, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2007-06-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.
J.Med.Chem., 50, 2007
|
|
2XTH
 
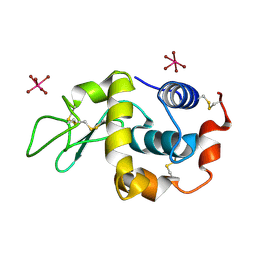 | | K2PtBr6 binding to lysozyme | | Descriptor: | HEXABROMOPLATINATE(IV), LYSOZYME C | | Authors: | Helliwell, J.R, Bell, A.M.T, Bryant, P, Fisher, S, Habash, J, Helliwell, M, Margiolaki, I, Kaenket, S, Watier, Y, Wright, J, Yalamanchili, S.K. | | Deposit date: | 2010-10-07 | | Release date: | 2010-12-08 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Time-Dependent Analysis of K2Ptbr6 Binding to Lysozyme Studied by Protein Powder and Single Crystal X-Ray Analysis
Z.Kristallogr., 225, 2010
|
|
2XWE
 
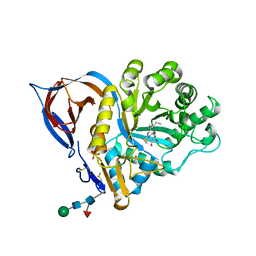 | | X-RAY STRUCTURE OF ACID-BETA-GLUCOSIDASE WITH 5N,6S-(N'-(N-OCTYL)IMINO)-6-THIONOJIRIMYCIN IN THE ACTIVE SITE | | Descriptor: | (3Z,5S,6R,7S,8R,8aS)-3-(octylimino)hexahydro[1,3]thiazolo[3,4-a]pyridine-5,6,7,8-tetrol, GLUCOSYLCERAMIDASE, PHOSPHATE ION, ... | | Authors: | Brumshtein, B, Aguilar-Moncayo, M, Benito, J.M, Ortiz Mellet, C, Garcia Fernandez, J.M, Silman, I, Shaaltiel, Y, Sussman, J.L, Futerman, A.H. | | Deposit date: | 2010-11-02 | | Release date: | 2011-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Cyclodextrin-Mediated Crystallization of Acid Beta-Glucosidase in Complex with Amphiphilic Bicyclic Nojirimycin Analogues.
Org.Biomol.Chem., 9, 2011
|
|
2XWD
 
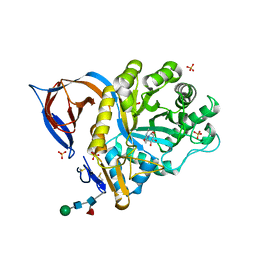 | | X-RAY STRUCTURE OF ACID-BETA-GLUCOSIDASE WITH 5N,6O-(N'-(N-OCTYL)IMINO)NOJIRIMYCIN IN THE ACTIVE SITE | | Descriptor: | (3Z,5S,6R,7S,8R,8aR)-3-(octylimino)hexahydro[1,3]oxazolo[3,4-a]pyridine-5,6,7,8-tetrol, GLUCOSYLCERAMIDASE, SULFATE ION, ... | | Authors: | Brumshtein, B, Aguilar-Moncayo, M, Benito, J.M, Ortiz Mellet, C, Garcia Fernandez, J.M, Silman, I, Shaaltiel, Y, Sussman, J.L, Futerman, A.H. | | Deposit date: | 2010-11-01 | | Release date: | 2011-09-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Cyclodextrin-Mediated Crystallization of Acid Beta-Glucosidase in Complex with Amphiphilic Bicyclic Nojirimycin Analogues.
Org.Biomol.Chem., 9, 2011
|
|
4ISV
 
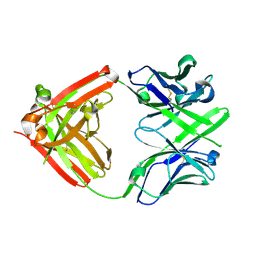 | | Crystal structure of the Fab FRAGMENT OF 1C2, A MONOCLONAL ANTIBODY SPECIFIC FOR POLY-GLUTAMINE | | Descriptor: | 1C2 FAB HEAVY CHAIN, 1C2 FAB LIGHT CHAIN | | Authors: | Klein, F.A.C, Zeder-Lutz, G, Cousido-Siah, A, Mitschler, A, Katz, A, Eberling, P, Mandel, J.L, Podjarny, A, Trottier, Y. | | Deposit date: | 2013-01-17 | | Release date: | 2013-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Linear and extended: a common polyglutamine conformation recognized by the three antibodies MW1, 1C2 and 3B5H10.
Hum.Mol.Genet., 22, 2013
|
|
2WCG
 
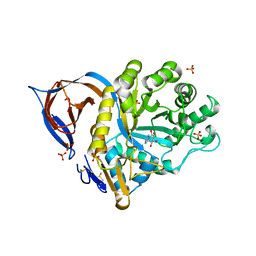 | | X-ray structure of acid-beta-glucosidase with N-octyl(cyclic guanidine)-nojirimycin in the active site | | Descriptor: | CHLORIDE ION, GLUCOSYLCERAMIDASE, N-[(3E,5R,6R,7S,8R,8AR)-5,6,7,8-TETRAHYDROXYHEXAHYDROIMIDAZO[1,5-A]PYRIDIN-3(2H)-YLIDENE]OCTAN-1-AMINIUM, ... | | Authors: | Brumshtein, B, Aguilar, M, Garcia-Moreno, M.I, Mellet, C.O, Garcia-Fernandez, J.M, Silman, I, Shaaltiel, Y, Aviezer, D, Sussman, J.L, Futerman, A.H. | | Deposit date: | 2009-03-12 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 6-Amino-6-Deoxy-5,6-Di-N-(N'-Octyliminomethylidene)Nojirimycin: Synthesis, Biological Evaluation, and Crystal Structure in Complex with Acid Beta-Glucosidase.
Chembiochem, 10, 2009
|
|
2V3E
 
 | | acid-beta-glucosidase with N-nonyl-deoxynojirimycin | | Descriptor: | (2R,3R,4R,5S)-2-(HYDROXYMETHYL)-1-NONYLPIPERIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUCOSYLCERAMIDASE, ... | | Authors: | Brumshtein, B, Greenblatt, H.M, Butters, T.D, Shaaltiel, Y, Aviezer, D, Silman, I, Futerman, A.H, Sussman, J.L. | | Deposit date: | 2007-06-17 | | Release date: | 2007-08-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Complexes of N-Butyl- and N-Nonyl-Deoxynojirimycin Bound to Acid Beta-Glucosidase: Insights Into the Mechanism of Chemical Chaperone Action in Gaucher Disease.
J.Biol.Chem., 282, 2007
|
|
4JJ5
 
 | | CRYSTAL STRUCTURE OF THE Fab FRAGMENT OF 1C2, A MONOCLONAL ANTIBODY SPECIFIC for POLY-GLUTAMINE | | Descriptor: | 1C2 FAB HEAVY CHAIN, 1C2 FAB LIGHT CHAIN | | Authors: | Klein, F.A.C, Zeder-Lutz, G, Cousido-Siah, A, Mitschler, A, Katz, A, Eberling, P, Mandel, J.L, Podjarny, A, Trottier, Y. | | Deposit date: | 2013-03-07 | | Release date: | 2013-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Linear and extended: a common polyglutamine conformation recognized by the three antibodies MW1, 1C2 and 3B5H10.
Hum.Mol.Genet., 22, 2013
|
|
2V3D
 
 | | acid-beta-glucosidase with N-butyl-deoxynojirimycin | | Descriptor: | (2R,3R,4R,5S)-1-BUTYL-2-(HYDROXYMETHYL)PIPERIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUCOSYLCERAMIDASE, ... | | Authors: | Brumshtein, B, Greenblatt, H.M, Butters, T.D, Shaaltiel, Y, Aviezer, D, Silman, I, Futerman, A.H, Sussman, J.L. | | Deposit date: | 2007-06-17 | | Release date: | 2007-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures of Complexes of N-Butyl- and N-Nonyl-Deoxynojirimycin Bound to Acid Beta-Glucosidase: Insights Into the Mechanism of Chemical Chaperone Action in Gaucher Disease.
J.Biol.Chem., 282, 2007
|
|
3VCM
 
 | | Crystal structure of human prorenin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, prorenin | | Authors: | Morales, R, Watier, Y, Bocskei, Z. | | Deposit date: | 2012-01-04 | | Release date: | 2012-05-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Human Prorenin Structure Sheds Light on a Novel Mechanism of Its Autoinhibition and on Its Non-Proteolytic Activation by the (Pro)renin Receptor.
J.Mol.Biol., 421, 2012
|
|
3S56
 
 | | HIV-1 protease triple mutants V32I, I47V, V82I with antiviral drug saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, ACETATE ION, Protease | | Authors: | Tie, Y.-F, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Critical differences in HIV-1 and HIV-2 protease specificity for clinical inhibitors.
Protein Sci., 21, 2012
|
|
3S45
 
 | | wild-type HIV-2 protease with antiviral drug amprenavir | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Protease, ... | | Authors: | Tie, Y.-F, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2011-05-18 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Critical differences in HIV-1 and HIV-2 protease specificity for clinical inhibitors.
Protein Sci., 21, 2012
|
|
3S53
 
 | | HIV-1 protease triple mutants V32I, I47V, V82I with antiviral drug darunavir in space group P212121 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, IODIDE ION, PHOSPHATE ION, ... | | Authors: | Tie, Y.-F, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Critical differences in HIV-1 and HIV-2 protease specificity for clinical inhibitors.
Protein Sci., 21, 2012
|
|
3S54
 
 | | HIV-1 protease triple mutants V32I, I47V, V82I with antiviral drug darunavir in space group P21212 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Tie, Y.-F, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Critical differences in HIV-1 and HIV-2 protease specificity for clinical inhibitors.
Protein Sci., 21, 2012
|
|
3S43
 
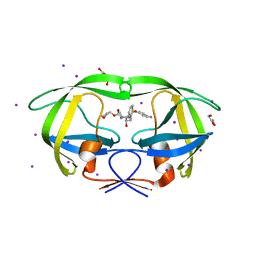 | | HIV-1 protease triple mutants V32I, I47V, V82I with antiviral drug amprenavir | | Descriptor: | GLYCEROL, IODIDE ION, Protease, ... | | Authors: | Wang, Y.-F, Tie, Y.-F, Weber, I.T. | | Deposit date: | 2011-05-18 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Critical differences in HIV-1 and HIV-2 protease specificity for clinical inhibitors.
Protein Sci., 21, 2012
|
|
2F8G
 
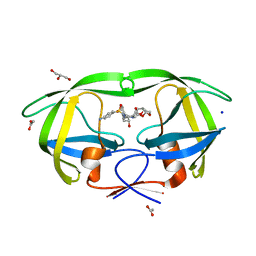 | | HIV-1 protease mutant I50V complexed with inhibitor TMC114 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2005-12-02 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Effectiveness of Nonpeptide Clinical Inhibitor TMC-114 on HIV-1 Protease with Highly Drug Resistant Mutations D30N, I50V, and L90M.
J.Med.Chem., 49, 2006
|
|
2F80
 
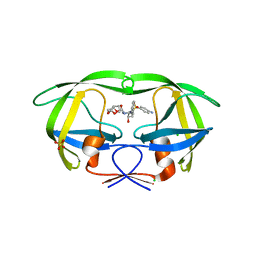 | | HIV-1 Protease mutant D30N complexed with inhibitor TMC114 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, POL POLYPROTEIN, ... | | Authors: | Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2005-12-01 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Effectiveness of Nonpeptide Clinical Inhibitor TMC-114 on HIV-1 Protease with Highly Drug Resistant Mutations D30N, I50V, and L90M.
J.Med.Chem., 49, 2006
|
|
2F81
 
 | | HIV-1 Protease mutant L90M complexed with inhibitor TMC114 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2005-12-01 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Effectiveness of Nonpeptide Clinical Inhibitor TMC-114 on HIV-1 Protease with Highly Drug Resistant Mutations D30N, I50V, and L90M.
J.Med.Chem., 49, 2006
|
|
4FL8
 
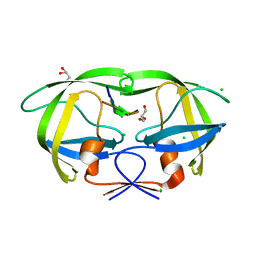 | | HIV-1 protease complexed with gem-diol-amine tetrahedral intermediate | | Descriptor: | CHLORIDE ION, GLYCEROL, HIV-1 protease, ... | | Authors: | Tie, Y.F, Shen, C.H, Weber, I.T. | | Deposit date: | 2012-06-14 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Capturing the Reaction Pathway in Near-Atomic-Resolution Crystal Structures of HIV-1 Protease.
Biochemistry, 51, 2012
|
|
3D1X
 
 | | Crystal structure of HIV-1 mutant I54M and inhibitor saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-05-06 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
3CYW
 
 | | Effect of Flap Mutations on Structure of HIV-1 Protease and Inhibition by Saquinavir and Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-04-27 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
4FLG
 
 | | HIV-1 protease mutant I47V complexed with reaction intermediate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Yu, X, Shen, C.H, Weber, I.T. | | Deposit date: | 2012-06-14 | | Release date: | 2012-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Capturing the Reaction Pathway in Near-Atomic-Resolution Crystal Structures of HIV-1 Protease.
Biochemistry, 51, 2012
|
|
4FM6
 
 | |
