5D6W
 
 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A | | Descriptor: | Lysine-specific demethylase 4A, S,R MESO-TARTARIC ACID | | Authors: | Wang, F, Su, Z, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
7R81
 
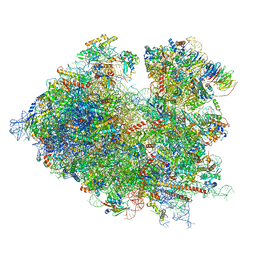 | | Structure of the translating Neurospora crassa ribosome arrested by cycloheximide | | Descriptor: | 18S rRNA, 26S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Shen, L, Su, Z, Yang, K, Wu, C, Becker, T, Bell-Pedersen, D, Zhang, J, Sachs, M.S. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the translating Neurospora ribosome arrested by cycloheximide
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7YR7
 
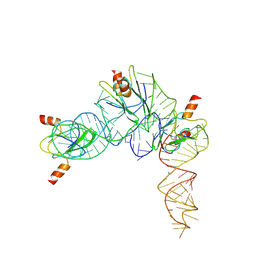 | | Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with three RsmA protein dimers | | Descriptor: | RsmZ RNA (118-MER), Translational regulator CsrA | | Authors: | Jia, X, Pan, Z, Yuan, Y, Luo, B, Luo, Y, Mukherjee, S, Jia, G, Liu, L, Ling, X, Yang, X, Wu, Y, Liu, T, Miao, Z, Wei, X, Bujnicki, J.M, Zhao, K, Su, Z. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of sRNA RsmZ regulation of Pseudomonas aeruginosa virulence.
Cell Res., 33, 2023
|
|
7YR6
 
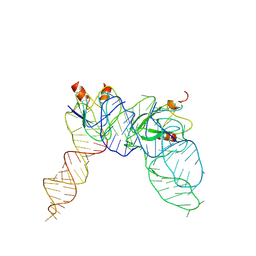 | | Cryo-EM structure of Pseudomonas aeruginosa RsmZ RNA in complex with two RsmA protein dimers | | Descriptor: | RsmZ RNA, Translational regulator CsrA | | Authors: | Jia, X, Pan, Z, Yuan, Y, Luo, B, Luo, Y, Mukherjee, S, Jia, G, Ling, X, Yang, X, Wu, Y, Liu, T, Wei, X, Bujnick, J.M, Zhao, K, Su, Z. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of sRNA RsmZ regulation of Pseudomonas aeruginosa virulence.
Cell Res., 33, 2023
|
|
7XD7
 
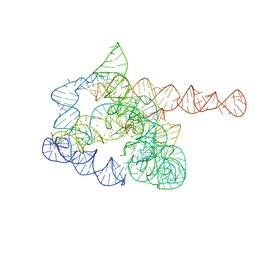 | | The pre-Tet-C state of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The pre-Tet-C state molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 30nt 3'/5'-exon | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD3
 
 | | The relaxed pre-Tet-S1 state of wild-type Tetrahymena group I intron with 6nt 3'/5'-exon | | Descriptor: | MAGNESIUM ION, The relaxed pre-Tet-S1 state molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 6nt 3'/5'-exon | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD4
 
 | | The intermediate pre-Tet-S1 state of wild-type Tetrahymena group I intron with 6nt 3'/5'-exon | | Descriptor: | Co-transcriptional folded wild-type Tetrahymena group I intron with 6nt 3'/5'-exon, MAGNESIUM ION | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD5
 
 | | The Tet-S2 state of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The Tet-S2 state molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 30nt 3'/5'-exon (5'-exon), ... | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD6
 
 | | The Tet-S2 state with a pseudoknotted 4-way junction of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The Tet-S2 state with a pseudoknotted 4-way junction molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 30nt 3'/5'-exon (5'-exon), ... | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
6P9E
 
 | | Crystal structure of IL-36gamma complexed to A-552 | | Descriptor: | (2S)-2-{[4-(3-amino-4-methylphenyl)-6-methylpyrimidin-2-yl]oxy}-3-methoxy-3,3-diphenylpropanoic acid, Interleukin-36 gamma | | Authors: | Argiriadi, M.A, Todorovic, T, Su, Z, Putman, B, Kakavas, S.J, Salte, K.M, McDonald, H.A, Wetter, J.B, Paulsboe, S.E, Sun, Q, Gerstein, C.E, Medina, L, Sielaff, B, Sadhukhan, R, Stockmann, H, Richardson, P.L, Qiu, W, Henry, R.F, Herold, J.M, Shotwell, J.B, McGaraughty, S.P, Honore, P, Gopalakrishnan, S.M, Sun, C.C, Scott, V.E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small Molecule IL-36 gamma Antagonist as a Novel Therapeutic Approach for Plaque Psoriasis.
Sci Rep, 9, 2019
|
|
6POM
 
 | | Cryo-EM structure of the full-length Bacillus subtilis glyQS T-box riboswitch in complex with tRNA-Gly | | Descriptor: | T-box GlyQS leader (155-MER), tRNAGly (75-MER) | | Authors: | Li, S, Su, Z, Zhang, J, Chiu, W. | | Deposit date: | 2019-07-04 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of amino acid surveillance by higher-order tRNA-mRNA interactions.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6WLM
 
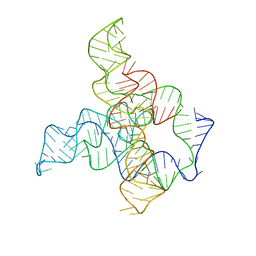 | | F. nucleatum glycine riboswitch with glycine models, 7.4 Angstrom resolution | | Descriptor: | RNA (171-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLR
 
 | | SAM-IV riboswitch with SAM models, 4.8 Angstrom resolution | | Descriptor: | RNA (119-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WQ2
 
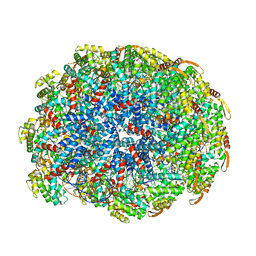 | | Cryo-EM of the S. islandicus filamentous virus, SIFV | | Descriptor: | A-DNA, Structural protein MCP1, Structural protein MCP2 | | Authors: | Wang, F, Baquero, D.P, Su, Z, Zheng, W, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of filamentous viruses infecting hyperthermophilic archaea explain DNA stabilization in extreme environments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WLS
 
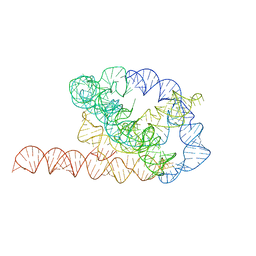 | | Tetrahymena ribozyme models, 6.8 Angstrom resolution | | Descriptor: | RNA (388-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
7Y5G
 
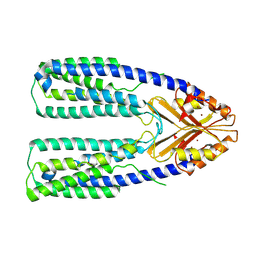 | | Cryo-EM structure of a eukaryotic ZnT8 in the presence of zinc | | Descriptor: | ZINC ION, Zinc transporter 8 | | Authors: | Zhang, S, Fu, C, Luo, Y, Sun, Z, Su, Z, Zhou, X. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Cryo-EM structure of a eukaryotic zinc transporter at a low pH suggests its Zn 2+ -releasing mechanism.
J.Struct.Biol., 215, 2022
|
|
7Y5H
 
 | | Cryo-EM structure of a eukaryotic ZnT8 at a low pH | | Descriptor: | ZINC ION, Zinc transporter 8 | | Authors: | Zhang, S, Fu, C, Luo, Y, Sun, Z, Su, Z, Zhou, X. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Cryo-EM structure of a eukaryotic zinc transporter at a low pH suggests its Zn 2+ -releasing mechanism.
J.Struct.Biol., 215, 2022
|
|
7XSN
 
 | | Native Tetrahymena ribozyme conformation | | Descriptor: | RNA (387-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSM
 
 | | Misfolded Tetrahymena ribozyme conformation 3 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSL
 
 | | Misfolded Tetrahymena ribozyme conformation 2 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSK
 
 | | Misfolded Tetrahymena ribozyme conformation 1 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7LJY
 
 | | Cryo-EM structure of the B dENE construct complexed with a 28-mer poly(A) | | Descriptor: | B dENE construct, poly(A) | | Authors: | Torabi, S, Chen, Y, Zhang, K, Wang, J, DeGregorio, S, Vaidya, A, Su, Z, Pabit, S, Chiu, W, Pollack, L, Steitz, J. | | Deposit date: | 2021-02-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural analyses of an RNA stability element interacting with poly(A).
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4HBN
 
 | | Crystal structure of the human HCN4 channel C-terminus carrying the S672R mutation | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, PHOSPHATE ION, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Xu, X, Marni, F, Wu, X, Su, Z, Musayev, F, Shrestha, S, Xie, C, Gao, W, Liu, Q, Zhou, L. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Local and Global Interpretations of a Disease-Causing Mutation near the Ligand Entry Path in Hyperpolarization-Activated cAMP-Gated Channel.
Structure, 20, 2012
|
|
8GQD
 
 | | Complex Structure of Arginine Kinase McsB and McsA from Staphylococcus aureus | | Descriptor: | Protein-arginine kinase, Protein-arginine kinase activator protein, ZINC ION | | Authors: | Lu, K, Luo, B, Tao, X, Li, H, Xie, Y, Zhao, Z, Xia, W, Su, Z, Mao, Z. | | Deposit date: | 2022-08-30 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Complex structure and activation mechanism of arginine kinase McsB by McsA.
Nat.Chem.Biol., 2024
|
|
8XIT
 
 | | Cryo-EM structure of sheep VMAT2 dimer in an atypical fold | | Descriptor: | OaVMAT2-BRIL | | Authors: | Lyu, Y, Fu, C, Ma, H, Sun, Z, Su, Z, Zhou, X. | | Deposit date: | 2023-12-20 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Engineering of a mammalian VMAT2 for cryo-EM analysis results in non-canonical protein folding.
Nat Commun, 15, 2024
|
|
