5T6M
 
 | | Structure of the tryptophan synthase b-subunit from Pyroccus furiosus with b-methyltryptophan non-covalently bound | | Descriptor: | (betaS)-beta-methyl-L-tryptophan, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Buller, A.R, van Roye, P, Arnold, F.H. | | Deposit date: | 2016-09-01 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tryptophan Synthase Uses an Atypical Mechanism To Achieve Substrate Specificity.
Biochemistry, 55, 2016
|
|
6AM7
 
 | |
6AM9
 
 | | Engineered tryptophan synthase b-subunit from Pyrococcus furiosus, PfTrpB2B9, with Ser-bound in a predominantly closed state. | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, SODIUM ION, Tryptophan synthase beta chain 1, ... | | Authors: | Buller, A.R, van Roye, P. | | Deposit date: | 2017-08-09 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Directed Evolution Mimics Allosteric Activation by Stepwise Tuning of the Conformational Ensemble.
J. Am. Chem. Soc., 140, 2018
|
|
6AM8
 
 | |
1MPP
 
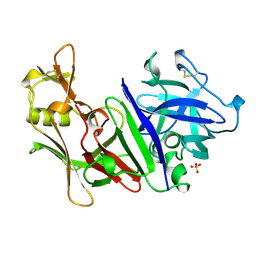 | | X-RAY ANALYSES OF ASPARTIC PROTEINASES. V. STRUCTURE AND REFINEMENT AT 2.0 ANGSTROMS RESOLUTION OF THE ASPARTIC PROTEINASE FROM MUCOR PUSILLUS | | Descriptor: | PEPSIN, SULFATE ION | | Authors: | Newman, M, Watson, F, Roychowdhury, P, Jones, H, Badasso, M, Cleasby, A, Wood, S.P, Tickle, I.J, Blundell, T.L. | | Deposit date: | 1992-02-19 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray analyses of aspartic proteinases. V. Structure and refinement at 2.0 A resolution of the aspartic proteinase from Mucor pusillus.
J.Mol.Biol., 230, 1993
|
|
8FU7
 
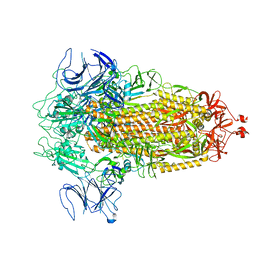 | | Structure of Covid Spike variant deltaN135 in fully closed form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
8FU8
 
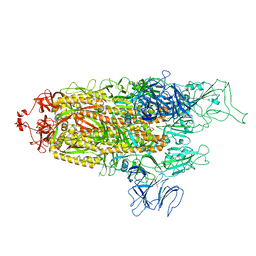 | | Structure of Covid Spike variant deltaN135 with one erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
8FU9
 
 | | Structure of Covid Spike variant deltaN25 with one erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
5G1L
 
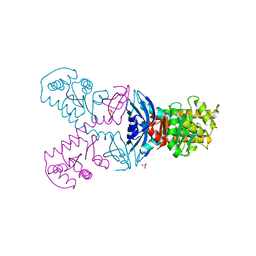 | | A double mutant of DsbG engineered for denitrosylation | | Descriptor: | SULFATE ION, THIOL DISULFIDE INTERCHANGE PROTEIN DSBG | | Authors: | Tamu Dufe, V, Van Molle, I, Lafaye, C, Wahni, K, Boudier, A, Leroy, P, Collet, J.F, Messens, J. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sulfur Denitrosylation by an Engineered Trx-Like Dsbg Enzyme Identifies Nucleophilic Cysteine Hydrogen Bonds as Key Functional Determinant.
J.Biol.Chem., 291, 2016
|
|
5G1K
 
 | | A triple mutant of DsbG engineered for denitrosylation | | Descriptor: | SULFATE ION, THIOL DISULFIDE INTERCHANGE PROTEIN DSBG | | Authors: | Tamu Dufe, V, Van Molle, I, Lafaye, C, Wahni, K, Boudier, A, Leroy, P, Collet, J.F, Messens, J. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Sulfur Denitrosylation by an Engineered Trx-Like Dsbg Enzyme Identifies Nucleophilic Cysteine Hydrogen Bonds as Key Functional Determinant.
J.Biol.Chem., 291, 2016
|
|
1AHS
 
 | |
1BVP
 
 | |
7NDS
 
 | | Crystal structure of TphC in a closed conformation | | Descriptor: | Tripartite tricarboxylate transporter substrate binding protein, terephthalic acid | | Authors: | Levy, C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of terephthalate recognition by solute binding protein TphC.
Nat Commun, 12, 2021
|
|
7NDR
 
 | | Crystal structure of TphC in an open conformation | | Descriptor: | 1,2-ETHANEDIOL, Tripartite tricarboxylate transporter substrate binding protein | | Authors: | Levy, C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of terephthalate recognition by solute binding protein TphC.
Nat Commun, 12, 2021
|
|
1A6S
 
 | | M-DOMAIN FROM GAG POLYPROTEIN OF ROUS SARCOMA VIRUS, NMR, 20 STRUCTURES | | Descriptor: | GAG POLYPROTEIN | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Zhou, W, Wolven, A, Wilson, C.B, Nelle, T.D, Resh, M.D, Wills, J, Cowburn, D. | | Deposit date: | 1998-03-02 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the bioactive retroviral M domain from Rous sarcoma virus
J.Mol.Biol., 279, 1998
|
|
8JFV
 
 | | Crystal structure of Catabolite repressor acivator from E. coli in complex with sulisobenzone | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-4-oxidanyl-5-(phenylcarbonyl)benzenesulfonic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Neetu, N, Sharma, M, Mahto, J.K, Kumar, P. | | Deposit date: | 2023-05-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Sulisobenzone is a potent inhibitor of the global transcription factor Cra.
J.Struct.Biol., 215, 2023
|
|
8JFF
 
 | |
1LQF
 
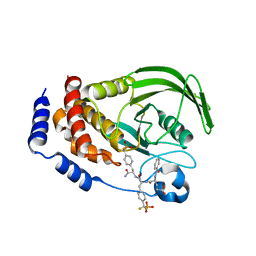 | | Structure of PTP1b in Complex with a Peptidic Bisphosphonate Inhibitor | | Descriptor: | N-BENZOYL-L-GLUTAMYL-[4-PHOSPHONO(DIFLUOROMETHYL)]-L-PHENYLALANINE-[4-PHOSPHONO(DIFLUORO-METHYL)]-L-PHENYLALANINEAMIDE, protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Asante-Appiah, E, Patel, S, Dufresne, C, Scapin, G. | | Deposit date: | 2002-05-10 | | Release date: | 2002-07-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of PTP-1B in complex with a peptide inhibitor reveals an alternative binding mode for bisphosphonates.
Biochemistry, 41, 2002
|
|
5IXJ
 
 | |
6AMH
 
 | |
6AMC
 
 | |
6AMI
 
 | |
8DZV
 
 | |
8VYE
 
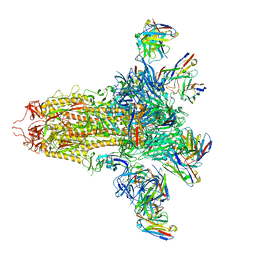 | | SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S2L20 Heavy Chain, S2L20 Light Chain, ... | | Authors: | McCallum, M, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-02-08 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Quantifying how single dose Ad26.COV2.S vaccine efficacy depends on Spike sequence features.
Nat Commun, 15, 2024
|
|
8VYG
 
 | | SARS-CoV-2 S RBD (C.37 Lambda variant) plus S309 Fab, local refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S309 Heavy Chain, S309 Light Chain, ... | | Authors: | McCallum, M, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-02-08 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Quantifying how single dose Ad26.COV2.S vaccine efficacy depends on Spike sequence features.
Nat Commun, 15, 2024
|
|
