1BKU
 
 | | EFFECTS OF GLYCOSYLATION ON THE STRUCTURE AND DYNAMICS OF EEL CALCITONIN, NMR, 10 STRUCTURES | | Descriptor: | CALCITONIN | | Authors: | Hashimoto, Y, Nishikido, J, Toma, K, Yamamoto, K, Haneda, K, Inazu, T, Valentine, K, Opella, S.J. | | Deposit date: | 1998-07-13 | | Release date: | 1999-01-13 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
1BYV
 
 | | GLYCOSYLATED EEL CALCITONIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CALCITONIN) | | Authors: | Hashimoto, Y, Toma, K, Nishikido, J, Yamamoto, K, Haneda, K, Inazu, T, Valentine, K.G, Opella, S.J. | | Deposit date: | 1998-10-16 | | Release date: | 1998-10-28 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
2KSJ
 
 | | Structure and Dynamics of the Membrane-bound form of Pf1 Coat Protein: Implications for Structural Rearrangement During Virus Assembly | | Descriptor: | Capsid protein G8P | | Authors: | Park, S, Marassi, F, Black, D, Opella, S.J. | | Deposit date: | 2010-01-05 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR, SOLUTION NMR | | Cite: | Structure and dynamics of the membrane-bound form of Pf1 coat protein: implications of structural rearrangement for virus assembly.
Biophys.J., 99, 2010
|
|
2LNL
 
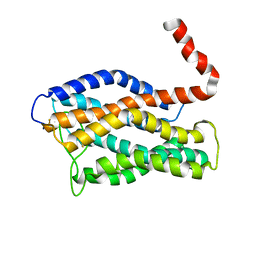 | | Structure of human CXCR1 in phospholipid bilayers | | Descriptor: | C-X-C chemokine receptor type 1 | | Authors: | Park, S, Das, B.B, Casagrande, F, Nothnagel, H, Chu, M, Kiefer, H, Maier, K, De Angelis, A, Marassi, F.M, Opella, S.J. | | Deposit date: | 2011-12-31 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-06 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the chemokine receptor CXCR1 in phospholipid bilayers.
Nature, 491, 2012
|
|
2LJ2
 
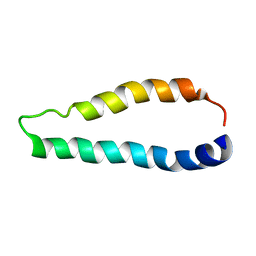 | | Integral membrane core domain of the mercury transporter MerF in lipid bilayer membranes | | Descriptor: | MerF | | Authors: | Das, B.B, Nothnagel, H.J, Lu, G.J, Son, W, Park, S, Tian, Y.B, Marassi, F.M, Opella, S.J. | | Deposit date: | 2011-09-03 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure determination of a membrane protein in proteoliposomes.
J.Am.Chem.Soc., 134, 2012
|
|
2MAG
 
 | | NMR STRUCTURE OF MAGAININ 2 IN DPC MICELLES, 10 STRUCTURES | | Descriptor: | MAGAININ 2 | | Authors: | Gesell, J.J, Zasloff, M, Opella, S.J. | | Deposit date: | 1997-12-19 | | Release date: | 1998-04-08 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Two-dimensional 1H NMR experiments show that the 23-residue magainin antibiotic peptide is an alpha-helix in dodecylphosphocholine micelles, sodium dodecylsulfate micelles, and trifluoroethanol/water solution.
J.Biomol.NMR, 9, 1997
|
|
2N28
 
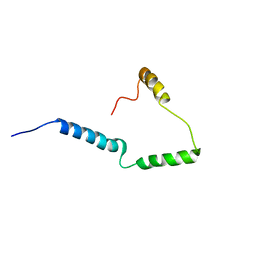 | | Solid-state NMR structure of Vpu | | Descriptor: | Protein Vpu | | Authors: | Zhang, H, Lin, E.C, Tian, Y, Das, B.B, Opella, S.J. | | Deposit date: | 2015-05-01 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Structural determination of virus protein U from HIV-1 by NMR in membrane environments.
Biochim.Biophys.Acta, 1848, 2015
|
|
2N29
 
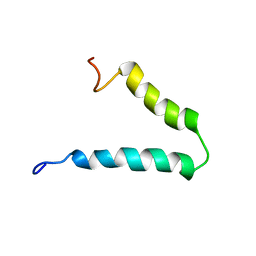 | | Solution-state NMR structure of Vpu cytoplasmic domain | | Descriptor: | Protein Vpu | | Authors: | Zhang, H, Lin, E.C, Tian, Y, Das, B.B, Opella, S.J. | | Deposit date: | 2015-05-01 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural determination of virus protein U from HIV-1 by NMR in membrane environments.
Biochim.Biophys.Acta, 1848, 2015
|
|
2MTS
 
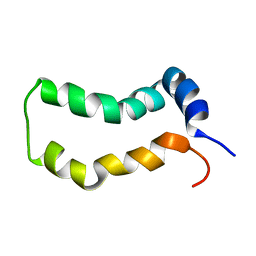 | | Three-Dimensional Structure and Interaction Studies of Hepatitis C Virus p7 in 1,2-Dihexanoyl-sn-glycero-3-phosphocholine by Solution Nuclear Magnetic Resonance | | Descriptor: | HEPATITIS C VIRUS P7 PROTEIN | | Authors: | Cook, G.A, Dawson, L.A, Tian, Y, Opella, S.J. | | Deposit date: | 2014-08-29 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and interaction studies of hepatitis C virus p7 in 1,2-dihexanoyl-sn-glycero-3-phosphocholine by solution nuclear magnetic resonance.
Biochemistry, 52, 2013
|
|
2KLV
 
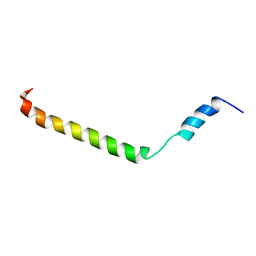 | | Membrane-bound structure of the Pf1 major coat protein in DHPC micelle | | Descriptor: | Capsid protein G8P | | Authors: | Park, S, Son, W, Mukhopadhyay, R, Valafar, H, Opella, S.J. | | Deposit date: | 2009-07-08 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Phage-induced alignment of membrane proteins enables the measurement and structural analysis of residual dipolar couplings with dipolar waves and lambda-maps.
J.Am.Chem.Soc., 131, 2009
|
|
2K3M
 
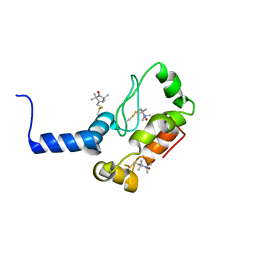 | | Rv1761c | | Descriptor: | Rv1761c, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Page, R.C, Moore, J.D, Lee, S, Opella, S.J, Cross, T.A. | | Deposit date: | 2008-05-14 | | Release date: | 2009-01-06 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Backbone structure of a small helical integral membrane protein: A unique structural characterization.
Protein Sci., 18, 2009
|
|
2MOZ
 
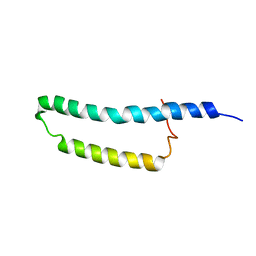 | | Structure of the Membrane Protein MerF, a Bacterial Mercury Transporter, Improved by the Inclusion of Chemical Shift Anisotropy Constraints | | Descriptor: | MerF | | Authors: | Tian, Y, Lu, G.J, Marassi, F.M, Opella, S.J, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2014-05-07 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the membrane protein MerF, a bacterial mercury transporter, improved by the inclusion of chemical shift anisotropy constraints.
J.Biomol.Nmr, 60, 2014
|
|
2M67
 
 | | Full-length mercury transporter protein MerF in lipid bilayer membranes | | Descriptor: | MerF | | Authors: | Lu, G.J, Tian, Y, Vora, N, Marassi, F.M, Opella, S.J. | | Deposit date: | 2013-03-27 | | Release date: | 2013-07-03 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The Structure of the Mercury Transporter MerF in Phospholipid Bilayers: A Large Conformational Rearrangement Results from N-Terminal Truncation.
J.Am.Chem.Soc., 135, 2013
|
|
2NR1
 
 | | TRANSMEMBRANE SEGMENT 2 OF NMDA RECEPTOR NR1, NMR, 10 STRUCTURES | | Descriptor: | NR1 M2 | | Authors: | Gesell, J.J, Sun, W, Montal, M, Opella, S. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy.
Nat.Struct.Biol., 6, 1999
|
|
4KEH
 
 | | Crosslinked Crystal Structure of Type II Fatty Synthase Dehydratase, FabA, and Acyl Carrier Protein, AcpP | | Descriptor: | Acyl carrier protein, N-{3-[DIHYDROXY(NONYL)-LAMBDA~4~-SULFANYL]PROPYL}-N~3~-[(2R)-2-HYDROXY-3,3-DIMETHYL-4-(PHOSPHONOOXY)BUTANOYL]-BETA-ALANINAMIDE, N-{3-[dihydroxy(nonyl)-lambda~4~-sulfanyl]propyl}-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide | | Authors: | Nguyen, C, Haushalter, R, Finzel, K, Leong, J, Le, B.C, Burkart, M, Tsai, S.C. | | Deposit date: | 2013-04-25 | | Release date: | 2013-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Trapping the dynamic acyl carrier protein in fatty acid biosynthesis.
Nature, 505, 2014
|
|
2LK9
 
 | | Structure of BST-2/Tetherin Transmembrane Domain | | Descriptor: | Bone marrow stromal antigen 2 | | Authors: | Skasko, M, Wang, Y, Tian, Y, Tokarev, A, Munguia, J, Ruiz, A, Stephens, E, Opella, S, Guatelli, J. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | HIV-1 Vpu Protein Antagonizes Innate Restriction Factor BST-2 via Lipid-embedded Helix-Helix Interactions.
J.Biol.Chem., 287, 2012
|
|
2MCW
 
 | | Solid-state NMR structure of piscidin 3 in aligned 3:1 phosphatidylcholine/phosphoglycerol lipid bilayers | | Descriptor: | Piscidin-3 | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2024-11-06 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
2N5H
 
 | | PltL-holo | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Peptidyl carrier protein PltL | | Authors: | Jaremko, M.J, Lee, D.J, Burkart, M.D. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and Substrate Sequestration in the Pyoluteorin Type II Peptidyl Carrier Protein PltL.
J.Am.Chem.Soc., 137, 2015
|
|
2N5I
 
 | | PltL-pyrrolyl | | Descriptor: | (3R)-3-hydroxy-2,2-dimethyl-4-oxo-4-{[3-oxo-3-({2-[(1H-pyrrol-2-ylcarbonyl)amino]ethyl}amino)propyl]amino}butyl dihydrogen phosphate, Peptidyl carrier protein PltL | | Authors: | Jaremko, M.J, Lee, D.J, Burkart, M.D. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and Substrate Sequestration in the Pyoluteorin Type II Peptidyl Carrier Protein PltL.
J.Am.Chem.Soc., 137, 2015
|
|
2MCV
 
 | | Solid-state NMR structure of piscidin 1 in aligned 1:1 phosphatidylethanolamine/phosphoglycerol lipid bilayers | | Descriptor: | Moronecidin | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Hayden, R.M, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-30 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
2MCU
 
 | | Solid-state NMR structure of piscidin 1 in aligned 3:1 phosphatidylcholine/phosphoglycerol lipid bilayers | | Descriptor: | Moronecidin | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2014-03-19 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
2MCX
 
 | | Solid-state NMR structure of piscidin 3 in aligned 1:1 phosphatidylethanolamine/phosphoglycerol lipid bilayers | | Descriptor: | Piscidin-3 | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Wieczorek, W.E, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-30 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
2XKM
 
 | | Consensus structure of Pf1 filamentous bacteriophage from X-ray fibre diffraction and solid-state NMR | | Descriptor: | CAPSID PROTEIN G8P | | Authors: | Straus, S.K, P Scott, W.R, Schwieters, C.D, Marvin, D.A. | | Deposit date: | 2010-07-09 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | FIBER DIFFRACTION (3.3 Å), SOLID-STATE NMR | | Cite: | Consensus Structure of Pf1 Filamentous Bacteriophage from X-Ray Fibre Diffraction and Solid-State NMR.
Eur.Biophys.J., 40, 2011
|
|
2C0X
 
 | | MOLECULAR STRUCTURE OF FD FILAMENTOUS BACTERIOPHAGE REFINED WITH RESPECT TO X-RAY FIBRE DIFFRACTION AND SOLID-STATE NMR DATA | | Descriptor: | COAT PROTEIN B | | Authors: | Marvin, D.A, Welsh, L.C, Symmons, M.F, Scott, W.R.P, Straus, S.K. | | Deposit date: | 2005-09-08 | | Release date: | 2005-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Molecular Structure of Fd (F1, M13) Filamentous Bacteriophage Refined with Respect to X-Ray Fibre Diffraction and Solid-State NMR Data Supports Specific Models of Phage Assembly at the Bacterial Membrane.
J.Mol.Biol., 355, 2006
|
|
