4FI6
 
 | | Kinetic Stabilization of transthyretin through covalent modification of K15 by 3-(5-(3,5-dichlorophenyl)-1,3,4-oxadiazol-2-yl)-benzenesulfonamide | | Descriptor: | 3-[5-(3,5-dichlorophenyl)-1,3,4-oxadiazol-2-yl]benzenesulfonyl fluoride, Transthyretin | | Authors: | Connelly, S, Grimster, N, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2012-06-08 | | Release date: | 2013-02-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Aromatic Sulfonyl Fluorides Covalently Kinetically Stabilize Transthyretin to Prevent Amyloidogenesis while Affording a Fluorescent Conjugate.
J.Am.Chem.Soc., 135, 2013
|
|
3CN0
 
 | |
3CN3
 
 | |
4FI8
 
 | | Kinetic Stabilization of transthyretin through covalent modification of K15 by 4-bromo-3-(5-(3,5-dichloro-4-hydroxyphenyl)-1,3,4-oxadiazol-2-yl)-benzenesulfonamide | | Descriptor: | 4-bromo-3-[5-(3,5-dichloro-4-hydroxyphenyl)-1,3,4-oxadiazol-2-yl]benzenesulfonyl fluoride, Transthyretin | | Authors: | Connelly, S, Grimster, N, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2012-06-08 | | Release date: | 2013-02-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Aromatic Sulfonyl Fluorides Covalently Kinetically Stabilize Transthyretin to Prevent Amyloidogenesis while Affording a Fluorescent Conjugate.
J.Am.Chem.Soc., 135, 2013
|
|
4FI7
 
 | | Kinetic Stabilization of transthyretin through covalent modification of K15 by 3-(5-(3,5-dichloro-4-hydroxyphenyl)-1,3,4-oxadiazol-2-yl)-benzenesulfonamide | | Descriptor: | 3-[5-(3,5-dichloro-4-hydroxyphenyl)-1,3,4-oxadiazol-2-yl]benzenesulfonyl fluoride, Transthyretin | | Authors: | Connelly, S, Grimster, N, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2012-06-08 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Aromatic Sulfonyl Fluorides Covalently Kinetically Stabilize Transthyretin to Prevent Amyloidogenesis while Affording a Fluorescent Conjugate.
J.Am.Chem.Soc., 135, 2013
|
|
3CN4
 
 | |
3P3U
 
 | | Human transthyretin (TTR) complexed with 5-(2-ethoxyphenyl)-3-(pyridin-4-yl)-1,2,4-oxadiazole | | Descriptor: | 4-[5-(2-ethoxyphenyl)-1,2,4-oxadiazol-3-yl]pyridine, Transthyretin | | Authors: | Connelly, S, Wilson, I.A. | | Deposit date: | 2010-10-05 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Potent kinetic stabilizers that prevent transthyretin-mediated cardiomyocyte proteotoxicity.
Sci Transl Med, 3, 2011
|
|
3ESP
 
 | | Human transthyretin (TTR) complexed with N-(3,5-Dibromo-4-hydroxyphenyl)-3,5-dimethyl-4-hydroxybenzamide | | Descriptor: | N-(3,5-dibromo-4-hydroxyphenyl)-4-hydroxy-3,5-dimethylbenzamide, Transthyretin | | Authors: | Connelly, S, Wilson, I.A. | | Deposit date: | 2008-10-06 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Toward optimization of the second aryl substructure common to transthyretin amyloidogenesis inhibitors using biochemical and structural studies.
J.Med.Chem., 52, 2009
|
|
3P3S
 
 | |
3TCT
 
 | | Structure of wild-type TTR in complex with tafamidis | | Descriptor: | 2-(3,5-dichlorophenyl)-1,3-benzoxazole-6-carboxylic acid, Transthyretin | | Authors: | Connelly, S, Kelly, J.W, Wilson, I.A. | | Deposit date: | 2011-08-09 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3CN2
 
 | |
3ESO
 
 | | Human transthyretin (TTR) complexed with N-(3,5-Dibromo-4-hydroxyphenyl)-2,5-dichlorobenzamide | | Descriptor: | 2,5-dichloro-N-(3,5-dibromo-4-hydroxyphenyl)benzamide, Transthyretin | | Authors: | Connelly, S, Wilson, I.A. | | Deposit date: | 2008-10-06 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Toward optimization of the second aryl substructure common to transthyretin amyloidogenesis inhibitors using biochemical and structural studies.
J.Med.Chem., 52, 2009
|
|
4HJS
 
 | |
4HIQ
 
 | | The Structure of V122I Mutant Transthyretin in Complex with AG10 | | Descriptor: | 3-[3-(3,5-dimethyl-1H-pyrazol-4-yl)propoxy]-4-fluorobenzoic acid, Transthyretin | | Authors: | Connelly, S, Alhamadsheh, M, Graef, I, Wilson, I.A. | | Deposit date: | 2012-10-11 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | AG10 inhibits amyloidogenesis and cellular toxicity of the familial amyloid cardiomyopathy-associated V122I transthyretin.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ESN
 
 | | Human transthyretin (TTR) complexed with N-(3,5-Dibromo-4-hydroxyphenyl)-2,6-dimethylbenzamide | | Descriptor: | N-(3,5-dibromo-4-hydroxyphenyl)-2,6-dimethylbenzamide, Transthyretin | | Authors: | Connelly, S, Wilson, I.A. | | Deposit date: | 2008-10-06 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Toward optimization of the second aryl substructure common to transthyretin amyloidogenesis inhibitors using biochemical and structural studies.
J.Med.Chem., 52, 2009
|
|
4HIS
 
 | | The Structure of V122I Mutant Transthyretin in Complex with Tafamidis | | Descriptor: | 2-(3,5-dichlorophenyl)-1,3-benzoxazole-6-carboxylic acid, Transthyretin | | Authors: | Connelly, S, Alhamadsheh, M, Graef, I, Wilson, I.A. | | Deposit date: | 2012-10-11 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | AG10 inhibits amyloidogenesis and cellular toxicity of the familial amyloid cardiomyopathy-associated V122I transthyretin.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HJU
 
 | |
7NAS
 
 | | Bacterial 30S ribosomal subunit assembly complex state A (multibody refinement for body domain of 30S ribosome) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7NAT
 
 | | Bacterial 30S ribosomal subunit assembly complex state A (Consensus refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7NAX
 
 | | Complete Bacterial 30S ribosomal subunit assembly complex state I (Consensus Refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7NAR
 
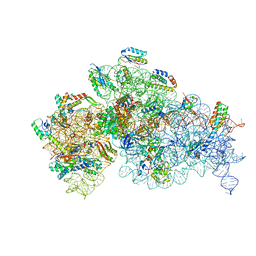 | | Complete Bacterial 30S ribosomal subunit assembly complex state F (+RsgA)(Consensus Refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7NAU
 
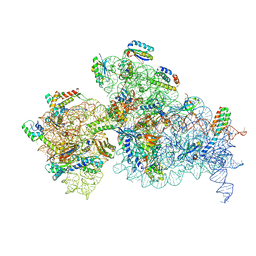 | | Bacterial 30S ribosomal subunit assembly complex state C (Consensus Refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7NAV
 
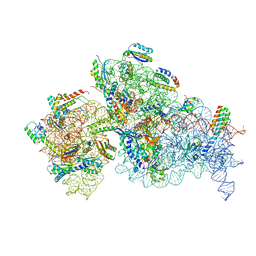 | | Bacterial 30S ribosomal subunit assembly complex state D (Consensus refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
5ME0
 
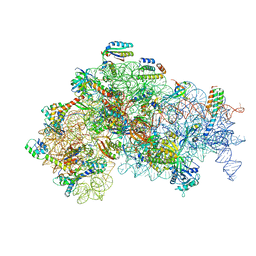 | | Structure of the 30S Pre-Initiation Complex 1 (30S IC-1) Stalled by GE81112 | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Fabbretti, A, Kaminishi, T, Iturrioz, I, Brandi, L, Gil Carton, D, Gualerzi, C, Fucini, P, Connell, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-01-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Structure of a 30S pre-initiation complex stalled by GE81112 reveals structural parallels in bacterial and eukaryotic protein synthesis initiation pathways.
Nucleic Acids Res., 45, 2017
|
|
5ME1
 
 | | Structure of the 30S Pre-Initiation Complex 2 (30S IC-2) Stalled by GE81112 | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Fabbretti, A, Kaminishi, T, Iturrioz, I, Brandi, L, Gil Carton, D, Gualerzi, C, Fucini, P, Connell, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-01-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Structure of a 30S pre-initiation complex stalled by GE81112 reveals structural parallels in bacterial and eukaryotic protein synthesis initiation pathways.
Nucleic Acids Res., 45, 2017
|
|
