7ZIF
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-480 | | Descriptor: | (2R)-2-azanyl-5-[[2-[3-methyl-2,6-bis(oxidanylidene)-7-(phenylmethyl)purin-8-yl]sulfanyl-3H-benzimidazol-5-yl]amino]-5-oxidanylidene-pentanoic acid, FE (III) ION, GLYCEROL, ... | | Authors: | Schuetz, A, Ziebart, N, Weise, M, Mallow, K, Pfeifer, J, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86859715 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
3C0Z
 
 | | Crystal structure of catalytic domain of human histone deacetylase HDAC7 in complex with SAHA | | Descriptor: | Histone deacetylase 7a, OCTANEDIOIC ACID HYDROXYAMIDE PHENYLAMIDE, POTASSIUM ION, ... | | Authors: | Min, J, Schuetz, A, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-21 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity.
J.Biol.Chem., 283, 2008
|
|
3C0Y
 
 | | Crystal structure of catalytic domain of human histone deacetylase HDAC7 | | Descriptor: | Histone deacetylase 7a, POTASSIUM ION, ZINC ION | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Loppnau, P, Kwiatkowski, N.P, Mazitschek, R, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-21 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity.
J.Biol.Chem., 283, 2008
|
|
3C10
 
 | | Crystal structure of catalytic domain of human histone deacetylase HDAC7 in complex with Trichostatin A (TSA) | | Descriptor: | Histone deacetylase 7a, POTASSIUM ION, TRICHOSTATIN A, ... | | Authors: | Min, J, Schuetz, A, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-21 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity.
J.Biol.Chem., 283, 2008
|
|
1Z4R
 
 | | Human GCN5 Acetyltransferase | | Descriptor: | ACETYL COENZYME *A, General control of amino acid synthesis protein 5-like 2 | | Authors: | Dong, A, Bernstein, G, Schuetz, A, Antoshenko, T, Wu, H, Loppnau, P, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Plotnikov, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-16 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of a binary complex between human GCN5 histone acetyltransferase domain and acetyl coenzyme A
Proteins, 68, 2007
|
|
2H9L
 
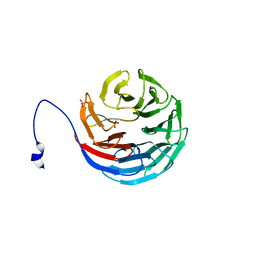 | | WDR5delta23 | | Descriptor: | SULFATE ION, WD-repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-10 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for molecular recognition and presentation of histone H3 By WDR5.
Embo J., 25, 2006
|
|
2H9P
 
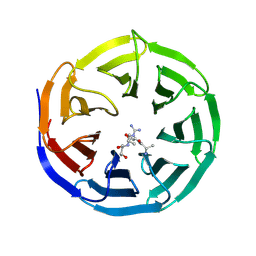 | | WDR5 in complex with trimethylated H3K4 peptide | | Descriptor: | H3 histone, WD-repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-10 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for molecular recognition and presentation of histone H3 By WDR5.
Embo J., 25, 2006
|
|
2H9M
 
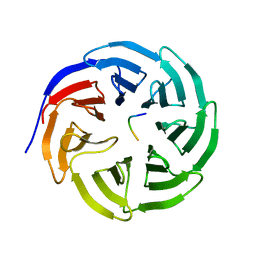 | | WDR5 in complex with unmodified H3K4 peptide | | Descriptor: | H3 histone, WD-repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-10 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for molecular recognition and presentation of histone H3 By WDR5.
Embo J., 25, 2006
|
|
2GNQ
 
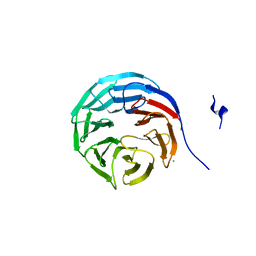 | | Structure of wdr5 | | Descriptor: | CHLORIDE ION, WD-repeat protein 5 | | Authors: | Min, J, Schuetz, A, Allali-Hassani, A, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-04-10 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for molecular recognition and presentation of histone H3 By WDR5.
Embo J., 25, 2006
|
|
2H9N
 
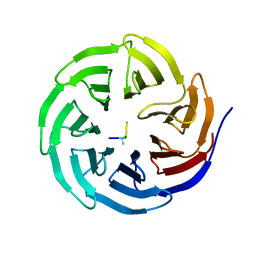 | | WDR5 in complex with monomethylated H3K4 peptide | | Descriptor: | H3 histone, WD-repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-10 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for molecular recognition and presentation of histone H3 By WDR5.
Embo J., 25, 2006
|
|
2O9K
 
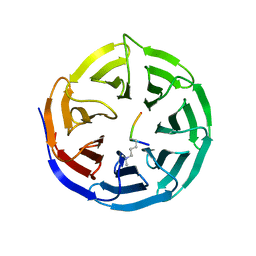 | | WDR5 in Complex with Dimethylated H3K4 Peptide | | Descriptor: | H3 HISTONE, WD repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-13 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Molecular Recognition and Presentation of Histone H3 by Wdr5.
Embo J., 25, 2006
|
|
2GL8
 
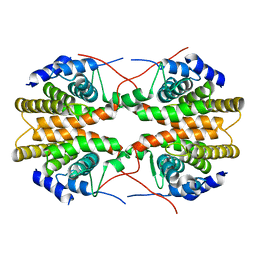 | | Human Retinoic acid receptor RXR-gamma ligand-binding domain | | Descriptor: | Retinoic acid receptor RXR-gamma | | Authors: | Min, J.R, Schuetz, A, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-04-04 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Human Retinoic acid receptor RXR-gamma ligand-binding domain.
To be Published
|
|
2PSW
 
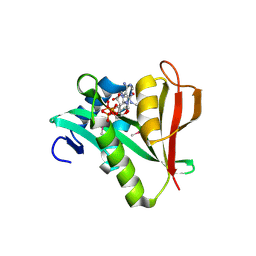 | | Human MAK3 homolog in complex with CoA | | Descriptor: | COENZYME A, N-acetyltransferase 13 | | Authors: | Walker, J.R, Schuetz, A, Antoshenko, T, Wu, H, Bernstein, G, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-07 | | Release date: | 2007-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Human MAK3 homolog.
To be Published
|
|
6EKU
 
 | | Vibrio cholerae neuraminidase complexed with zanamivir | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schuetz, A, Heinemann, U. | | Deposit date: | 2017-09-27 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Different Inhibitory Potencies of Oseltamivir Carboxylate, Zanamivir, and Several Tannins on Bacterial and Viral Neuraminidases as Assessed in a Cell-Free Fluorescence-Based Enzyme Inhibition Assay.
Molecules, 22, 2017
|
|
5MR7
 
 | |
6EKS
 
 | | Vibrio cholerae neuraminidase complexed with oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Schuetz, A, Heinemann, U. | | Deposit date: | 2017-09-27 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Different Inhibitory Potencies of Oseltamivir Carboxylate, Zanamivir, and Several Tannins on Bacterial and Viral Neuraminidases as Assessed in a Cell-Free Fluorescence-Based Enzyme Inhibition Assay.
Molecules, 22, 2017
|
|
3REA
 
 | |
3REB
 
 | |
2WBS
 
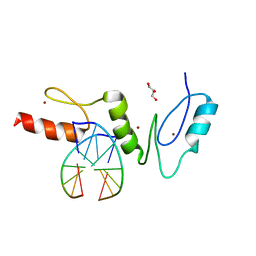 | | Crystal structure of the zinc finger domain of Klf4 bound to its target DNA | | Descriptor: | 5'-D(*GP*AP*GP*GP*CP*GP*CP)-3', 5'-D(*GP*CP*GP*CP*CP*TP*CP)-3', GLYCEROL, ... | | Authors: | Zocher, G, Schuetz, A, Carstanjen, D, Heinemann, U. | | Deposit date: | 2009-03-03 | | Release date: | 2010-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of the Klf4 DNA-Binding Domain Links to Self-Renewal and Macrophage Differentiation.
Cell.Mol.Life Sci., 68, 2011
|
|
3TT9
 
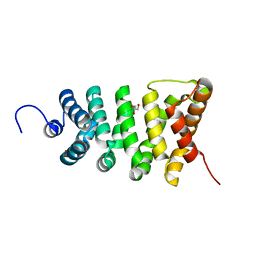 | | Crystal structure of the stable degradation fragment of human plakophilin 2 isoform a (PKP2a) C752R variant | | Descriptor: | GLYCEROL, Plakophilin-2 | | Authors: | Schuetz, A, Roske, Y, Gerull, B, Heinemann, U. | | Deposit date: | 2011-09-14 | | Release date: | 2012-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular insights into arrhythmogenic right ventricular cardiomyopathy caused by plakophilin-2 missense mutations.
Circ Cardiovasc Genet, 5, 2012
|
|
1H4V
 
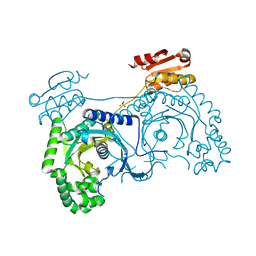 | |
1H3E
 
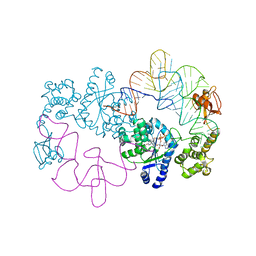 | | Tyrosyl-tRNA synthetase from Thermus thermophilus complexed with wild-type tRNAtyr(GUA) and with ATP and tyrosinol | | Descriptor: | 4-[(2S)-2-amino-3-hydroxypropyl]phenol, ADENOSINE-5'-TRIPHOSPHATE, TYROSYL-TRNA SYNTHETASE, ... | | Authors: | Cusack, S, Yaremchuk, A, Kriklivyi, I, Tukalo, M. | | Deposit date: | 2002-08-28 | | Release date: | 2002-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Class I Tyrosyl-tRNA Synthetase Has a Class II Mode or tRNA Recognition
Embo J., 21, 2002
|
|
1H3F
 
 | | Tyrosyl-tRNA synthetase from Thermus thermophilus complexed with tyrosinol | | Descriptor: | 4-[(2S)-2-amino-3-hydroxypropyl]phenol, SULFATE ION, TYROSYL-TRNA SYNTHETASE | | Authors: | Cusack, S, Yaremchuk, A, Kriklivyi, I, Tukalo, M. | | Deposit date: | 2002-08-28 | | Release date: | 2002-09-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Class I Tyrosyl-tRNA Synthetase Has a Class II Mode or tRNA Recognition
Embo J., 21, 2002
|
|
7NSG
 
 | | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B | | Descriptor: | (+)-3-Hydroxy-4,5,6,6a-tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-6-carboxylic acid, (-)-3-Hydroxy-4,5,6,6a-tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-6-carboxylic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Baronina, A, Pike, A.C.W, Yu, X, Dong, Y.Y, Shintre, C.A, Tessitore, A, Chu, A, Rotty, B, Venkaya, S, Mukhopadhyay, S.M.M, Borkowska, O, Chalk, R, Shrestha, L, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Han, S, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-05 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B
TO BE PUBLISHED
|
|
5LEV
 
 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) (V264G mutant) | | Descriptor: | UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase, UNKNOWN LIGAND | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
