9DCG
 
 | | Crystal Structure of the Thiol:Disulfide Interchange Protein DsbC from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Shatsman, S, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-08-26 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of the Thiol:Disulfide Interchange Protein DsbC from Vibrio cholerae
To Be Published
|
|
9DDY
 
 | | The Crystal Structure of Geranyltranstransferase from Streptococcus pneumoniae TIGR4 | | Descriptor: | GLYCEROL, Geranyltranstransferase | | Authors: | Kim, Y, Tan, A, Endres, M, Tan, K, Joachimik, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-08-28 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The Crystal Structure of Geranyltranstransferase from Streptococcus pneumoniae TIGR4
To Be Published
|
|
9MTO
 
 | | Crystal structure of acetyltransferase CBU0801 from Coxiella burnetii | | Descriptor: | COENZYME A, [Ribosomal protein bS18]-alanine N-acetyltransferase | | Authors: | Stogios, P.J, Evdokimova, E, Chang, C, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2025-01-11 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of acetyltransferase CBU0801 from Coxiella burnetii
To Be Published
|
|
8T28
 
 | | The crystal structure of SrtC2 sortase from Actinomyces oris | | Descriptor: | CHLORIDE ION, Class C sortase, PHOSPHATE ION | | Authors: | Osipiuk, J, Chang, C, Ton-That, H.L, Ton-That, H, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-06-05 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for dual functions in pilus assembly modulated by the lid of a pilus-specific sortase.
J.Biol.Chem., 300, 2024
|
|
8TTP
 
 | | Crystal structure of class C beta-lactamase from Escherichia coli in complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-08-14 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of class C beta-lactamase from Escherichia coli in complex with avibactam
to be published
|
|
8UFM
 
 | | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2 | | Descriptor: | ACETATE ION, FORMIC ACID, Papain-like protease nsp3, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-04 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2
To Be Published
|
|
8UFL
 
 | | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus | | Descriptor: | CHLORIDE ION, Papain-like protease nsp3, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-04 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus
To Be Published
|
|
8VLC
 
 | | Crystal structure of Zn-dependent hydrolase from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, HARLDQ motif MBL-fold protein, SULFATE ION | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-01-11 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Zn-dependent hydrolase from Salmonella typhimurium LT2
To Be Published
|
|
8SCD
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 in complex with reaction intermediate | | Descriptor: | 2-amino-6-methylidene-6,7-dihydropteridin-4(3H)-one, 4-AMINOBENZOIC ACID, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-05 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
8SMQ
 
 | | Crystal Structure of the N-terminal Domain of the Cryptic Surface Protein (CD630_25440) from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein target highlights in CASP15: Analysis of models by structure providers.
Proteins, 91, 2023
|
|
7UUJ
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with gentamicin | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, ... | | Authors: | Stogios, P.J, Evdokimova, E, Osipiuk, J, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2023-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with gentamicin
To Be Published
|
|
7UUK
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with tobramycin | | Descriptor: | Aminocyclitol acetyltransferase ApmA, CHLORIDE ION, TOBRAMYCIN | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
8U00
 
 | | Crystal structure of metallo-beta-lactamase superfamily protein from Caulobacter vibrioides | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-08-28 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of metallo-beta-lactamase superfamily protein from Caulobacter vibrioides
To Be Published
|
|
8U12
 
 | |
8UFB
 
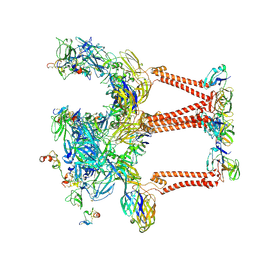 | |
8UFC
 
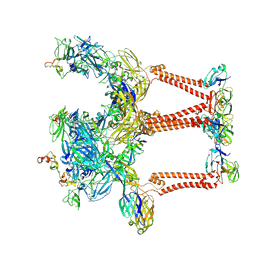 | |
8UFA
 
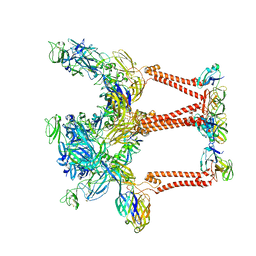 | |
9P0P
 
 | | Crystal structure of 23S rRNA methyltransferase Cfr, apoenzyme | | Descriptor: | Ribosomal RNA large subunit methyltransferase Cfr | | Authors: | Stogios, P.J, Skarina, T, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2025-06-07 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 23S rRNA methyltransferase Cfr, apoenzyme
To Be Published
|
|
9OZ0
 
 | | Crystal structure of T6SS effector-immunity complex PA3907-PA3908 from Pseudomonas aeruginosa | | Descriptor: | CITRIC ACID, Immunity protein 52 domain-containing protein, MAGNESIUM ION, ... | | Authors: | Stogios, P.J, Borek, D, Skarina, T, Osipiuk, J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2025-06-04 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.871 Å) | | Cite: | Crystal structure of T6SS effector-immunity complex PA3907-PA3908 from Pseudomonas aeruginosa
To Be Published
|
|
8UQU
 
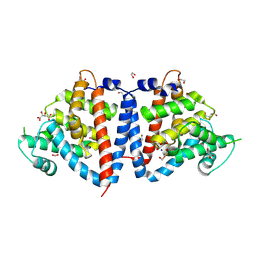 | | Crystal Structure of N-terminal Domain of Fic Family Protein from Bordetella bronchiseptica | | Descriptor: | 1,2-ETHANEDIOL, D(-)-TARTARIC ACID, Fido domain-containing protein, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-24 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of N-terminal Domain of Fic Family Protein from Bordetella bronchiseptica
To Be Published
|
|
8UW6
 
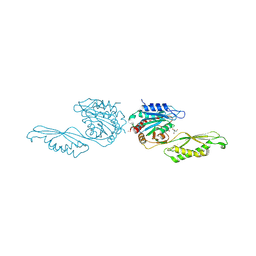 | | Acetylornithine deacetylase from Escherichia coli, di-zinc form. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acetylornithine deacetylase, ... | | Authors: | Osipiuk, J, Endres, M, Kelley, E, Becker, D.P, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-11-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | N alpha-acetyl-L-ornithine deacetylase from Escherichia coli and a ninhydrin-based assay to enable inhibitor identification.
Front Chem, 12, 2024
|
|
8UND
 
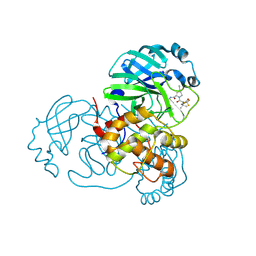 | | X-ray Structure of SARS-CoV-2 main protease covalently bound to inhibitor GRL-190-21 at 1.90 A. | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopiperidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, ORF1a polyprotein | | Authors: | Mesecar, A.D, Lendy, E.K, Ghosh, A.K, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-18 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray Structure of SARS-CoV-2 main protease covalently bound to inhibitor GRL-190-21 at 1.90 A.
To Be Published
|
|
7SWW
 
 | |
7SWX
 
 | |
7SUA
 
 | | Crystal Structure of the Hypothetical Protein (ACX60_00475) from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DUF4175 domain-containing protein | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the Hypothetical Protein (ACX60_00475) from Acinetobacter baumannii
To be Published
|
|
