4UT9
 
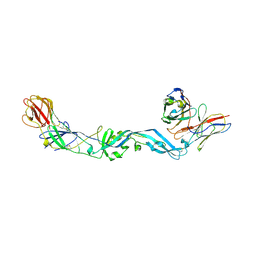 | | Crystal structure of dengue 2 virus envelope glycoprotein dimer in complex with the ScFv fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C10, ENVELOPE GLYCOPROTEIN E | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
4UTA
 
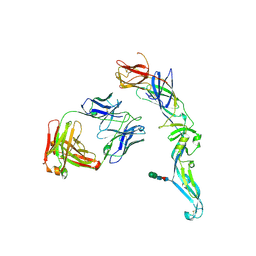 | | Crystal structure of dengue 2 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE1 C8 | | Descriptor: | BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C8 HEAVY CHAIN, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C8 LIGHT CHAIN, ENVELOPE GLYCOPROTEIN E, ... | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
4V1L
 
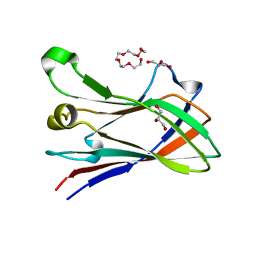 | | High resolution structure of a novel carbohydrate binding module from glycoside hydrolase family 9 (Cel9A) from Ruminococcus flavefaciens FD-1 | | Descriptor: | CARBOHYDRATE BINDING MODULE, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Venditto, I, Goyal, A, Thompson, A, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4UX6
 
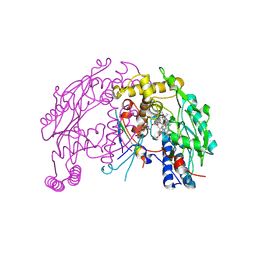 | | The discovery of novel, potent and highly selective inhibitors of inducible nitric oxide synthase (iNOS) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, NITRIC OXIDE SYNTHASE, INDUCIBLE, ... | | Authors: | Cheshire, D.R, Andrews, G, Beaton, H.G, Birkinshaw, T, Boughton-Smith, N, Connolly, S, Cook, T.R, Cooper, A, Cooper, S.L, Cox, D, Dixon, J, Gensmantel, N, Hamley, P.J, Harrison, R, Hartopp, P, Kack, H, Luker, T, Mete, A, Millichip, I, Nicholls, D.J, Pimm, A.D, St-Gallay, S.A, Wallace, A.V. | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-08 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Discovery of Novel, Potent and Highly Selective Inhibitors of Inducible Nitric Oxide Synthase (Inos).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7X56
 
 | | A CBg-ParM filament with ADP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ParM/StbA family protein | | Authors: | Koh, A, Ali, S, Robinson, R, Narita, A. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A new actin-like filament from Clostridium botulinum exhibits a new system of filament turn over
To Be Published
|
|
7X59
 
 | | A CBg-ParM filament with GTP or GDPPi | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, ParM/StbA family protein | | Authors: | Koh, A, Ali, S, Robinson, R, Narita, A. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | A new actin-like filament from Clostridium botulinum exhibits a new system of filament turn over
To Be Published
|
|
7X55
 
 | |
7X54
 
 | | A CBg-ParM filament with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ParM/StbA family protein | | Authors: | Koh, A, Ali, S, Robinson, R, Narita, A. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A new actin-like filament from Clostridium botulinum exhibits a new system of filament turn over
To Be Published
|
|
7XBJ
 
 | | Txp40, an insecticidal toxin protein from Xenorhabdus nematophila | | Descriptor: | 40kDa insecticidal toxin | | Authors: | Kinkar, O, Kumar, A, Prashar, A, Hire, R.S, Makde, R.D. | | Deposit date: | 2022-03-21 | | Release date: | 2023-03-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The crystal structure of insecticidal protein Txp40 from Xenorhabdus nematophila reveals a two-domain unique binary toxin with homology to the toxin-antitoxin (TA) system.
Insect Biochem.Mol.Biol., 164, 2023
|
|
7XMP
 
 | |
4WU7
 
 | | HLA-A24 in complex with HIV-1 Nef134-8(2F) | | Descriptor: | 8-Mer peptide from Protein Nef, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shimizu, A, Yamagata, A, Fukai, S, Iwamoto, A. | | Deposit date: | 2014-10-31 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structure of Nef134-8 epitope presented by HLA-A24
To Be Published
|
|
4WBQ
 
 | | Crystal structure of the exonuclease domain of QIP (QDE-2 interacting protein) solved by native-SAD phasing. | | Descriptor: | CALCIUM ION, QDE-2-interacting protein | | Authors: | Boland, A, Weinert, T, Weichenrieder, O, Wang, M. | | Deposit date: | 2014-09-03 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
7A01
 
 | | The Halastavi arva virus intergenic region IRES promotes translation by the simplest possible initiation mechanism | | Descriptor: | 18S RIBOSOMAL RNA, 28S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN ES21, ... | | Authors: | Abaeva, I, Vicens, Q, Bochler, A, Soufari, H, Simonetti, A, Pestova, T.V, Hashem, Y, Hellen, C.U.T. | | Deposit date: | 2020-08-05 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The Halastavi arva Virus Intergenic Region IRES Promotes Translation by the Simplest Possible Initiation Mechanism.
Cell Rep, 33, 2020
|
|
6ZOK
 
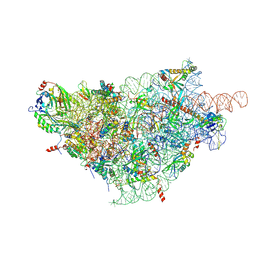 | | SARS-CoV-2-Nsp1-40S complex, focused on body | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M, Thiel, V, Muehlemann, O, Ban, N. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZSD
 
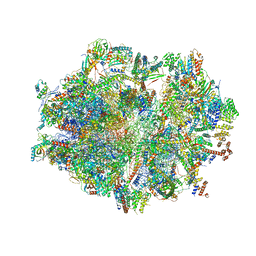 | | Human mitochondrial ribosome in complex with mRNA, P-site tRNA and E-site tRNA | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Aibara, S, Singh, V, Modelska, A, Amunts, A. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of mitochondrial translation.
Elife, 9, 2020
|
|
7X5F
 
 | | Nrf2-MafG heterodimer bound with CsMBE2 | | Descriptor: | Nuclear factor erythroid 2-related factor 2, Synthetic DNA, Transcription factor MafG | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
7XAR
 
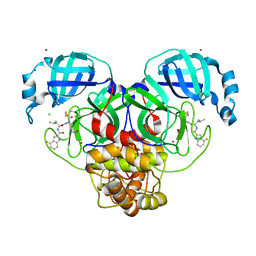 | | Crystal structure of 3C-like protease from SARS-CoV-2 in complex with covalent inhibitor | | Descriptor: | 3C-like proteinase, 4-fluoranyl-~{N}-[(2~{S})-1-[2-(2-fluoranylethanoyl)-2-[[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]methyl]hydrazinyl]-4-methyl-1-oxidanylidene-pentan-2-yl]-1~{H}-indole-2-carboxamide, CHLORIDE ION, ... | | Authors: | Caaveiro, J.M.M, Ochi, J, Takahashi, D, Ueda, T, Ojida, A. | | Deposit date: | 2022-03-18 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Chlorofluoroacetamide-Based Covalent Inhibitors for Severe Acute Respiratory Syndrome Coronavirus 2 3CL Protease.
J.Med.Chem., 65, 2022
|
|
7X5G
 
 | | Nrf2 (A510Y)-MafG heterodimer bound with CsMBE2 | | Descriptor: | DNA (5'-D(*CP*AP*CP*AP*GP*TP*GP*AP*CP*TP*CP*AP*GP*CP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*TP*GP*AP*GP*TP*CP*AP*CP*TP*GP*T)-3'), Nuclear factor erythroid 2-related factor 2, ... | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
7X5E
 
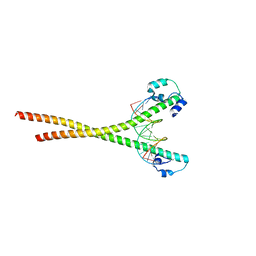 | | Nrf2-MafG heterodimer bound with CsMBE1 | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*TP*GP*AP*GP*TP*CP*AP*GP*CP*AP*A)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP*TP*CP*AP*T)-3'), HEXAETHYLENE GLYCOL, ... | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
6ZOJ
 
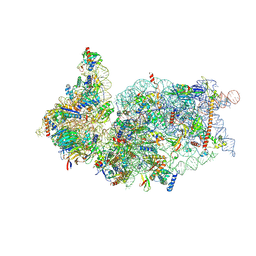 | | SARS-CoV-2-Nsp1-40S complex, composite map | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M.L, Thiel, V, Muehlemann, O, Ban, N. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-22 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4WZV
 
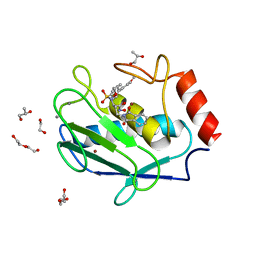 | | Crystal structure of a hydroxamate based inhibitor EN140 in complex with the MMP-9 catalytic domain | | Descriptor: | (2R)-4-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-N-hydroxy-2-{[(4'-methoxybiphenyl-4-yl)sulfonyl](propan-2-yloxy)amino}butanamide, 1,2-ETHANEDIOL, AZIDE ION, ... | | Authors: | Stura, E.A, Vera, L, Cassar-Lajeunesse, E, Nuti, E, Dive, V, Rossello, A. | | Deposit date: | 2014-11-20 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | N-O-Isopropyl Sulfonamido-Based Hydroxamates as Matrix Metalloproteinase Inhibitors: Hit Selection and in Vivo Antiangiogenic Activity.
J.Med.Chem., 58, 2015
|
|
6ZOL
 
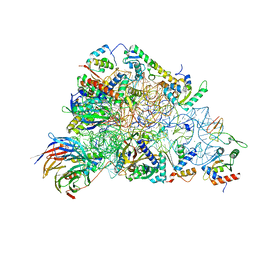 | | SARS-CoV-2-Nsp1-40S complex, focused on head | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S12, ... | | Authors: | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M.L, Thiel, V, Muehlemann, O, Ban, N. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5WCF
 
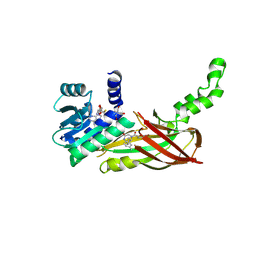 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | (5R)-4-(5-bromofuran-2-carbonyl)-5-(4-fluorophenyl)-7-methyl-1,3,4,5-tetrahydro-2H-1,4-benzodiazepin-2-one, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Dong, A, Zeng, H, Hutch, A, Seitova, A, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Crystal Structure of Human HMT1 hnRNP methyltransferase-like protein 6 in complex with MTLLE1441
to be published
|
|
6ZSB
 
 | | Human mitochondrial ribosome in complex with mRNA and P-site tRNA | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Aibara, S, Singh, V, Modelska, A, Amunts, A. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of mitochondrial translation.
Elife, 9, 2020
|
|
7A5V
 
 | | CryoEM structure of a human gamma-aminobutyric acid receptor, the GABA(A)R-beta3 homopentamer, in complex with histamine and megabody Mb25 in lipid nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nakane, T, Kotecha, A, Sente, A, Yamashita, K, McMullan, G, Masiulis, S, Brown, P.M.G.E, Grigoras, I.T, Malinauskaite, L, Malinauskas, T, Miehling, J, Yu, L, Karia, D, Pechnikova, E.V, de Jong, E, Keizer, J, Bischoff, M, McCormack, J, Tiemeijer, P, Hardwick, S.W, Chirgadze, D.Y, Murshudov, G, Aricescu, A.R, Scheres, S.H.W. | | Deposit date: | 2020-08-22 | | Release date: | 2020-11-18 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (1.7 Å) | | Cite: | Single-particle cryo-EM at atomic resolution.
Nature, 587, 2020
|
|
