6O9H
 
 | | Mouse ECD with Fab1 | | Descriptor: | Gastric inhibitory polypeptide receptor, Heavy chain, Light chain, ... | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-22 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism of an antagonistic antibody against glucose-dependent insulinotropic polypeptide receptor.
Mabs, 12, 2020
|
|
8HNW
 
 | | Crystal structure of HpaCas9-sgRNA surveillance complex bound to double-stranded DNA | | Descriptor: | CRISPR-associated endonuclease Cas9, Non-target strand, Target strand, ... | | Authors: | Sun, W, Cheng, Z, Wang, Y. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HNT
 
 | |
6O9I
 
 | | Ternary complex of mouse ECD with Fab1 and Fab2 | | Descriptor: | 1,2-ETHANEDIOL, Fab 2 heavy chain, Fab1 heavy chain, ... | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism of an antagonistic antibody against glucose-dependent insulinotropic polypeptide receptor.
Mabs, 12, 2020
|
|
8DOL
 
 | | Mechanism of regulation of the Helicobacter pylori Cagbeta ATPase by CagZ | | Descriptor: | Cag pathogenicity island protein (Cag5), DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Wu, X, Zhao, Y, Yang, W, Sun, L, Ye, X, Jiang, M, Wang, Q, Wang, Q, Zhang, X, Wu, Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of regulation of the Helicobacter pylori Cag beta ATPase by CagZ.
Nat Commun, 14, 2023
|
|
7BVP
 
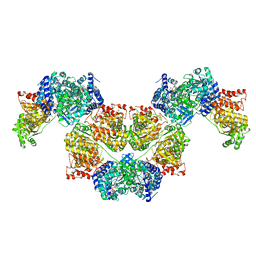 | | AdhE spirosome in extended conformation | | Descriptor: | Aldehyde-alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Kim, G.J, Song, J.J. | | Deposit date: | 2020-04-11 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Aldehyde-alcohol dehydrogenase undergoes structural transition to form extended spirosomes for substrate channeling.
Commun Biol, 3, 2020
|
|
6DXL
 
 | | Linked amidobenzimidazole STING agonist | | Descriptor: | 1,1'-(butane-1,4-diyl)bis{2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-1H-benzimidazole-5-carboxamide}, CALCIUM ION, Stimulator of interferon protein | | Authors: | Concha, N.O. | | Deposit date: | 2018-06-29 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design of amidobenzimidazole STING receptor agonists with systemic activity.
Nature, 564, 2018
|
|
6DXG
 
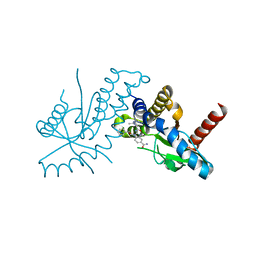 | | amidobenzimidazole (ABZI) STING agonists | | Descriptor: | 2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-1-[(2R)-2-hydroxy-2-phenylethyl]-1H-benzimidazole-5-carboxamide, CALCIUM ION, Stimulator of interferon protein | | Authors: | Concha, N.O. | | Deposit date: | 2018-06-28 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Design of amidobenzimidazole STING receptor agonists with systemic activity.
Nature, 564, 2018
|
|
5ZNT
 
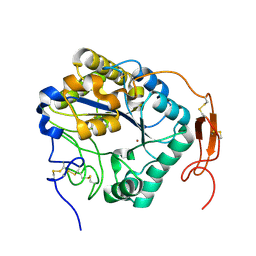 | | Insect chitin deacetylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ZINC ION, ... | | Authors: | Liu, L, Zhou, Y, Yang, Q. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structural and biochemical insights into the catalytic mechanisms of two insect chitin deacetylases of the carbohydrate esterase 4 family.
J. Biol. Chem., 294, 2019
|
|
5ZNS
 
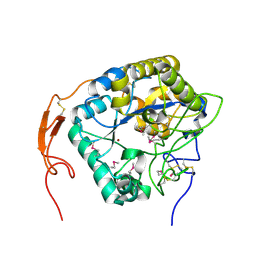 | | Insect chitin deacetylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ZINC ION, ... | | Authors: | Liu, L, Zhou, Y, Yang, Q. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structural and biochemical insights into the catalytic mechanisms of two insect chitin deacetylases of the carbohydrate esterase 4 family.
J. Biol. Chem., 294, 2019
|
|
3ADI
 
 | |
3ADL
 
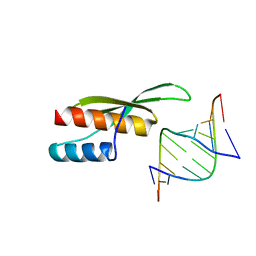 | |
8GYB
 
 | |
8GYA
 
 | |
8GY4
 
 | | Crystal structure of Alongshan virus methyltransferase | | Descriptor: | Methyltransferase | | Authors: | Chen, H, Lin, S, Lu, G.W. | | Deposit date: | 2022-09-21 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional basis of low-affinity SAM/SAH-binding in the conserved MTase of the multi-segmented Alongshan virus distantly related to canonical unsegmented flaviviruses.
Plos Pathog., 19, 2023
|
|
8GY9
 
 | |
3I3T
 
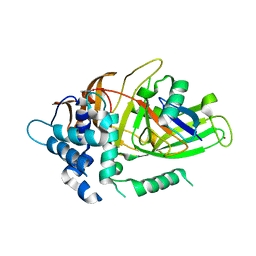 | | Crystal structure of covalent ubiquitin-USP21 complex | | Descriptor: | ETHANAMINE, Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 21, ... | | Authors: | Neculai, D, Avvakumov, G.V, Walker, J.R, Xue, S, Butler-Cole, C, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
8XIF
 
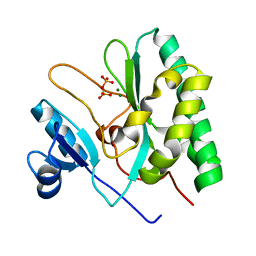 | | The crystal structure of the AEP domain of VACV D5 | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE, Uncoating factor OPG117 | | Authors: | Gan, J, Zhang, W. | | Deposit date: | 2023-12-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
8XJ8
 
 | | The Cryo-EM structure of MPXV E5 C-terminal in complex with DNA | | Descriptor: | DNA (70-MER), MAGNESIUM ION, Monkeypox virus E5, ... | | Authors: | Zhang, W, Liu, Y, Gao, H, Gan, J. | | Deposit date: | 2023-12-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
8XIG
 
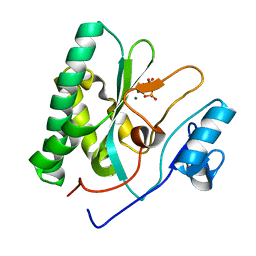 | | The crystal structure of the AEP domain of MPXV E5 | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE, Uncoating factor OPG117 | | Authors: | Gan, J, Zhang, W. | | Deposit date: | 2023-12-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
8XJ7
 
 | | The Cryo-EM structure of MPXV E5 in complex with DNA | | Descriptor: | DNA (70-MER), MAGNESIUM ION, Monkeypox virus E5, ... | | Authors: | Zhang, W, Liu, Y, Gao, H, Gan, J. | | Deposit date: | 2023-12-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
8XJ6
 
 | | The Cryo-EM structure of MPXV E5 apo conformation | | Descriptor: | AMP PHOSPHORAMIDATE, Monkeypox virus E5, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Zhang, W, Liu, Y, Gao, H, Gan, J. | | Deposit date: | 2023-12-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
6BS2
 
 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 8b | | Descriptor: | 1-(3,6-dimethyl[1,2]oxazolo[5,4-d]pyrimidin-4-yl)-6-methoxy-1,2,3,4-tetrahydroquinoline, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-12-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
6BR1
 
 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 4a | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-4-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)pyrido[2,3-d]pyrimidine, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
6BRF
 
 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 4b | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-4-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)pyrido[3,2-d]pyrimidine, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-11-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
