6AKR
 
 | | Crystal structure of the PDE4D catalytic domain in complex with osthole | | Descriptor: | 7-methoxy-8-(3-methylbut-2-enyl)chromen-2-one, ZINC ION, cAMP-specific 3',5'-cyclic phosphodiesterase 4D | | Authors: | Wang, S, Huo, Y.W, Xie, Y. | | Deposit date: | 2018-09-03 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.326 Å) | | Cite: | Airway relaxation mechanisms and structural basis of osthole for improving lung function in asthma.
Sci.Signal., 13, 2020
|
|
6K05
 
 | | Crystal structure of BRD2(BD1)with ligand BY27 bound | | Descriptor: | (6~{R})-~{N}-(4-chlorophenyl)-1-methyl-8-(1-methylpyrazol-4-yl)-5,6-dihydro-4~{H}-[1,2,4]triazolo[4,3-a][1]benzazepin-6-amine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2 | | Authors: | Lu, T, Lu, W, Chen, D, Zhao, Y, Luo, C. | | Deposit date: | 2019-05-05 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Discovery, structural insight, and bioactivities of BY27 as a selective inhibitor of the second bromodomains of BET proteins.
Eur.J.Med.Chem., 182, 2019
|
|
6K04
 
 | | Crystal structure of BRD2(BD2)with ligand BY27 bound | | Descriptor: | (6~{R})-~{N}-(4-chlorophenyl)-1-methyl-8-(1-methylpyrazol-4-yl)-5,6-dihydro-4~{H}-[1,2,4]triazolo[4,3-a][1]benzazepin-6-amine, Bromodomain-containing protein 2 | | Authors: | Lu, T, Lu, W, Chen, D, Zhao, Y, Luo, C. | | Deposit date: | 2019-05-05 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Discovery, structural insight, and bioactivities of BY27 as a selective inhibitor of the second bromodomains of BET proteins.
Eur.J.Med.Chem., 182, 2019
|
|
6IR1
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM4 at 1.9 Angstron resolution | | Descriptor: | MCherry fluorescent protein, mCherry's nanobody LaM4 | | Authors: | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | Deposit date: | 2018-11-09 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
7D3D
 
 | | Crystal structure of SPOP bound with a peptide | | Descriptor: | GLU-VAL-SER-ILE-ILE-GLN-GLY-ALA-ASP-SER-THR-THR, GLYCEROL, Speckle-type POZ protein | | Authors: | Yang, C.-G, Gan, J.H. | | Deposit date: | 2020-09-18 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A peptide binder of E3 ligase adaptor SPOP disrupts oncogenic SPOP-protein interactions in kidney cancer cells.
Chin.J.Chem., 39, 2021
|
|
7E6R
 
 | | Crystal structure of HCoV-NL63 3C-like protease,pH5.6 | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-23 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7E6L
 
 | | Crystal structure of HCoV-NL63 3C-like protease,pH5.0 | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78037143 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7E6M
 
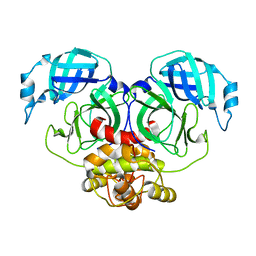 | | Crystal structure of Human coronavirus NL63 3C-like protease | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83445024 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7E6N
 
 | | Crystal structure of HCoV-NL63 3C-like protease,pH5.2 | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8413 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
6K7P
 
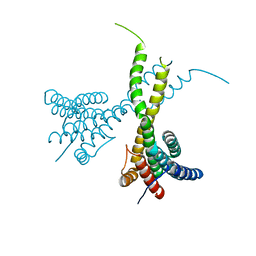 | | Crystal structure of human AFF4-THD domain | | Descriptor: | AF4/FMR2 family member 4 | | Authors: | Tang, D, Xue, Y, Li, S, Cheng, W, Duan, J, Wang, J, Qi, S. | | Deposit date: | 2019-06-08 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insight into the effect of AFF4 dimerization on activation of HIV-1 proviral transcription.
Cell Discov, 6, 2020
|
|
8IG4
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with GC376 | | Descriptor: | Non-structural protein 11, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IG5
 
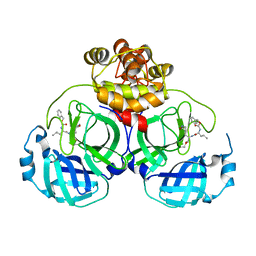 | | Crystal structure of SARS main protease in complex with GC376 | | Descriptor: | 3C-like proteinase nsp5, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IG6
 
 | | Crystal structure of MERS main protease in complex with GC376 | | Descriptor: | N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, ORF1a | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IG8
 
 | |
8IG7
 
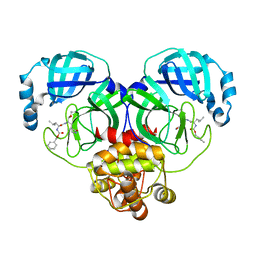 | |
8IG9
 
 | |
8IGB
 
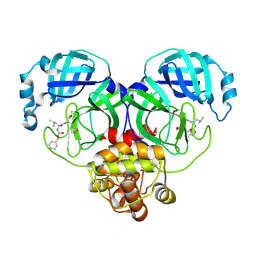 | |
8IGA
 
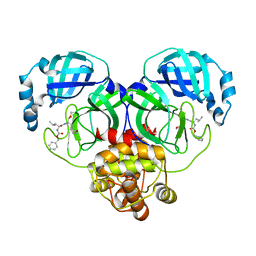 | |
8J37
 
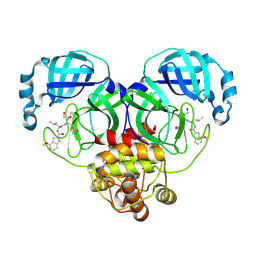 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by PF-00835231.
Acta Biochim.Biophys.Sin., 56, 2024
|
|
8J35
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by PF-00835231.
Acta Biochim.Biophys.Sin., 56, 2024
|
|
8J3A
 
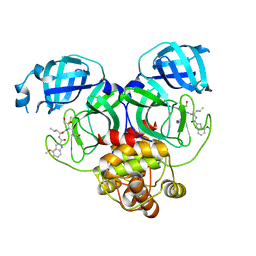 | | Crystal structure of SARS-Cov-2 main protease Y54C mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by PF-00835231.
Acta Biochim.Biophys.Sin., 56, 2024
|
|
8J32
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by PF-00835231.
Acta Biochim.Biophys.Sin., 56, 2024
|
|
8J38
 
 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by PF-00835231.
Acta Biochim.Biophys.Sin., 56, 2024
|
|
8J34
 
 | | Crystal structure of MERS main protease in complex with PF00835231 | | Descriptor: | N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ORF1a | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by PF-00835231.
Acta Biochim.Biophys.Sin., 56, 2024
|
|
8J3B
 
 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with PF00835231 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by PF-00835231.
Acta Biochim.Biophys.Sin., 56, 2024
|
|
