4OEX
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 6-ethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Ren, J, Xu, Y.C. | | Deposit date: | 2014-01-14 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Thermodynamic and structural characterization of halogen bonding in protein-ligand interactions: a case study of PDE5 and its inhibitors.
J.Med.Chem., 57, 2014
|
|
8XV2
 
 | | Thiamine-bound human SLC19A3 | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, Soluble cytochrome b562,Thiamine transporter 2 | | Authors: | Dang, Y, Wang, G.P, Zhang, Z. | | Deposit date: | 2024-01-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Substrate and drug recognition mechanisms of SLC19A3.
Cell Res., 34, 2024
|
|
8XV5
 
 | | Pyridoxamine-bound human SLC19A3 | | Descriptor: | 4-(AMINOMETHYL)-5-(HYDROXYMETHYL)-2-METHYLPYRIDIN-3-OL, Soluble cytochrome b562,Thiamine transporter 2 | | Authors: | Dang, Y, Wang, G.P, Zhang, Z. | | Deposit date: | 2024-01-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Substrate and drug recognition mechanisms of SLC19A3.
Cell Res., 34, 2024
|
|
6K0Y
 
 | | Study of the interactions of a novel monoclonal antibody, mAb059c, with the hPD-1 receptor | | Descriptor: | 1,2-ETHANEDIOL, Antibody Heavy Chain, Antibody Light Chain, ... | | Authors: | Liu, J.X, Wang, G.Q. | | Deposit date: | 2019-05-08 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Study of the interactions of a novel monoclonal antibody, mAb059c, with the hPD-1 receptor.
Sci Rep, 9, 2019
|
|
3F9V
 
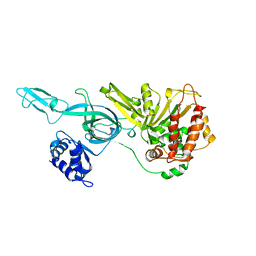 | | Crystal Structure Of A Near Full-Length Archaeal MCM: Functional Insights For An AAA+ Hexameric Helicase | | Descriptor: | Minichromosome maintenance protein MCM | | Authors: | Chen, X.J, Brewster, A.S, Wang, G.G, Yu, X, Greenleaf, W, Tjajadi, M, Klein, M. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Crystal structure of a near-full-length archaeal MCM: Functional insights for an AAA+ hexameric helicase.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
5DDV
 
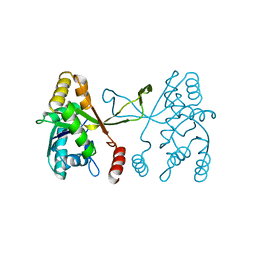 | |
7KGY
 
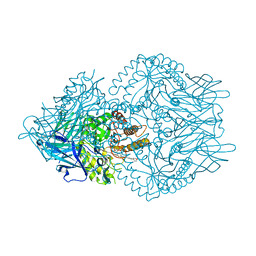 | | Beta-glucuronidase from Faecalibacterium prausnitzii bound to the inhibitor UNC10201652-glucuronide | | Descriptor: | 8-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-5-(morpholin-4-yl)-1,2,3,4-tetrahydro[1,2,3]triazino[4',5':4,5]thieno[2,3 -c]isoquinoline, Beta-glucuronidase, GLYCEROL | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2020-10-19 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Microbial enzymes induce colitis by reactivating triclosan in the mouse gastrointestinal tract.
Nat Commun, 13, 2022
|
|
7KGZ
 
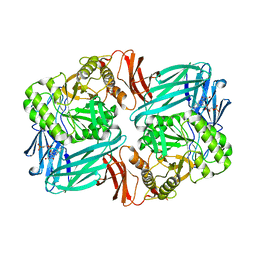 | |
5ZLM
 
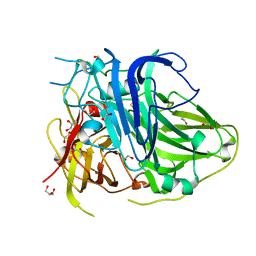 | | Mutation in the trinuclear site of CotA-laccase: H491C mutant, PH 8.0 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Xie, T, Liu, Z.C, Wang, G.G. | | Deposit date: | 2018-03-28 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insight into the Allosteric Coupling of Cu1 Site and Trinuclear Cu Cluster in CotA Laccase.
Chembiochem, 19, 2018
|
|
5ZLK
 
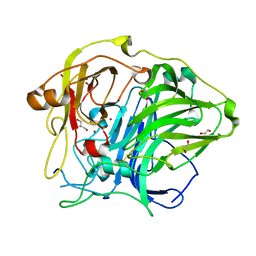 | | Mutation in the trinuclear site of CotA-laccase: H493A mutant, PH 8.0 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Xie, T, Liu, Z.C, Wang, G.G. | | Deposit date: | 2018-03-28 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insight into the Allosteric Coupling of Cu1 Site and Trinuclear Cu Cluster in CotA Laccase.
Chembiochem, 19, 2018
|
|
5X8H
 
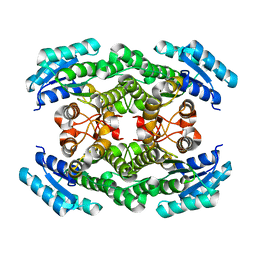 | | Crystal structure of the ketone reductase ChKRED20 from the genome of Chryseobacterium sp. CA49 | | Descriptor: | Short-chain dehydrogenase reductase | | Authors: | Zhao, F.J, Jin, Y, Liu, Z.C, Wang, G.G, Wu, Z.L. | | Deposit date: | 2017-03-02 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and iterative saturation mutagenesis of ChKRED20 for expanded catalytic scope
Appl. Microbiol. Biotechnol., 101, 2017
|
|
5ZLL
 
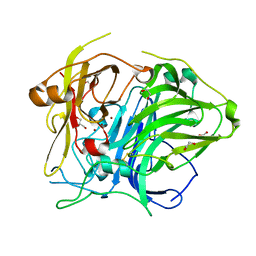 | | Mutation in the trinuclear site of CotA-laccase: H493C mutant, PH 8.0 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Xie, T, Liu, Z.C, Wang, G.G. | | Deposit date: | 2018-03-28 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insight into the Allosteric Coupling of Cu1 Site and Trinuclear Cu Cluster in CotA Laccase.
Chembiochem, 19, 2018
|
|
7CHF
 
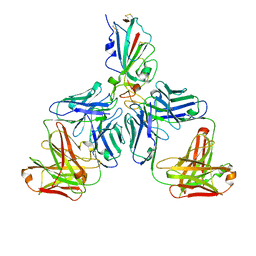 | |
7CHE
 
 | |
7CH4
 
 | |
7CHB
 
 | | Crystal structure of the SARS-CoV-2 RBD in complex with BD-236 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD-236 Fab heavy chain, BD-236 Fab light chain, ... | | Authors: | Xiao, J, Zhu, Q. | | Deposit date: | 2020-07-05 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy.
Cell, 183, 2020
|
|
8HGH
 
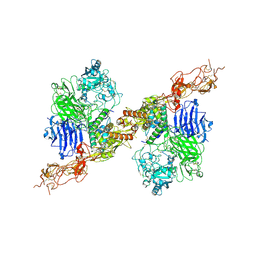 | | Structure of 2:2 PAPP-A.STC2 complex | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Pappalysin-1, Stanniocalcin-2, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
8HGG
 
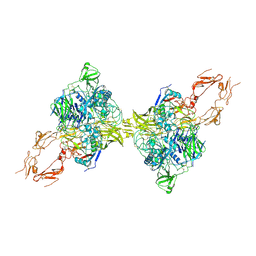 | | Structure of 2:2 PAPP-A.ProMBP complex | | Descriptor: | Bone marrow proteoglycan, Pappalysin-1, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
6IXM
 
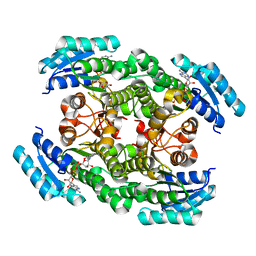 | | Crystal structure of the ketone reductase ChKRED20 from the genome of Chryseobacterium sp. CA49 complexed with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Short-chain dehydrogenase reductase | | Authors: | Zhao, F.J, Jin, Y, Liu, Z.C, Wang, G.G, Wu, Z.L. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structure-guided engineering of ChKRED20 from Chryseobacterium sp. CA49 for asymmetric reduction of aryl ketoesters.
Enzyme Microb. Technol., 125, 2019
|
|
7CHC
 
 | |
7CH5
 
 | |
6KLI
 
 | | Crystal Structure of the Zea Mays laccase 3 complexed with sinapyl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2,6-dimethoxyphenol, ... | | Authors: | Xie, T, Liu, Z.C, Wang, G.G. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for monolignol oxidation by a maize laccase.
Nat.Plants, 6, 2020
|
|
6KLJ
 
 | | Crystal Structure of the Zea Mays laccase 3 complexed with coniferyl | | Descriptor: | (2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enal, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xie, T, Liu, Z.C, Wang, G.G. | | Deposit date: | 2019-07-30 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural basis for monolignol oxidation by a maize laccase.
Nat.Plants, 6, 2020
|
|
6DNU
 
 | |
6DNL
 
 | | Crystal Structure of Neisseria meningitidis DsbD c-terminal domain in the reduced form | | Descriptor: | ACETATE ION, Thiol:disulfide interchange protein DsbD, ZINC ION | | Authors: | Smith, R.P, Heras, B, Paxman, J.J. | | Deposit date: | 2018-06-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical insights into the disulfide reductase mechanism of DsbD, an essential enzyme for neisserial pathogens.
J. Biol. Chem., 293, 2018
|
|
