3WY2
 
 | | Crystal structure of alpha-glucosidase in complex with glucose | | Descriptor: | Alpha-glucosidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shen, X, Gai, Z, Kato, K, Yao, M. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Structural analysis of the alpha-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
7XP6
 
 | | Cryo-EM structure of a class T GPCR in active state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
7XP4
 
 | | Cryo-EM structure of a class T GPCR in apo state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
7XP5
 
 | | Cryo-EM structure of a class T GPCR in ligand-free state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
6MC1
 
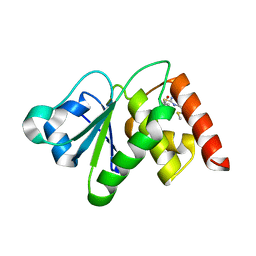 | | Structure of MAP kinase phosphatase 5 in complex with 3,3-dimethyl-1-((9-(methylthio)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl)thio)butan-2-one, an allosteric inhibitor | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3,3-dimethyl-1-{[9-(methylsulfanyl)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl]sulfanyl}butan-2-one, ACETATE ION, ... | | Authors: | Gannam, Z.T.K, Anderson, K.S, Bennett, A.M, Lolis, E. | | Deposit date: | 2018-08-30 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An allosteric site on MKP5 reveals a strategy for small-molecule inhibition.
Sci.Signal., 13, 2020
|
|
3WY1
 
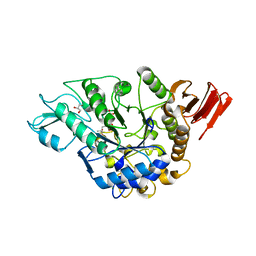 | | Crystal structure of alpha-glucosidase | | Descriptor: | (3R,5R,7R)-octane-1,3,5,7-tetracarboxylic acid, Alpha-glucosidase, GLYCEROL, ... | | Authors: | Shen, X, Gai, Z, Kato, K, Yao, M. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of the alpha-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
8JT8
 
 | | Crystal structure of 5-HT2AR in complex with (R)-IHCH-7179 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(4-fluorophenyl)-4-[(7R)-2,5,11-triazatetracyclo[7.6.1.0^2,7.0^12,16]hexadeca-1(15),9,12(16),13-tetraen-5-yl]butan-1-one, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Chen, Z, Fan, L, Wang, S. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Flexible scaffold-based cheminformatics approach for polypharmacological drug design.
Cell, 187, 2024
|
|
8JT6
 
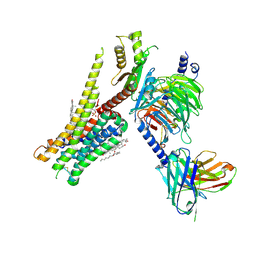 | | 5-HT1A-Gi in complex with compound (R)-IHCH-7179 | | Descriptor: | 1-(4-fluorophenyl)-4-[(7R)-2,5,11-triazatetracyclo[7.6.1.0^2,7.0^12,16]hexadeca-1(15),9,12(16),13-tetraen-5-yl]butan-1-one, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Chen, Z, Xu, P, Huang, S, Xu, H.E, Wang, S. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Flexible scaffold-based cheminformatics approach for polypharmacological drug design.
Cell, 187, 2024
|
|
5H06
 
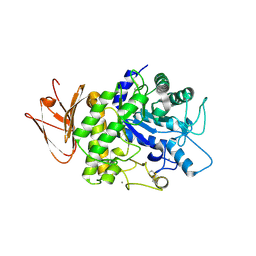 | | Crystal structure of AmyP in complex with maltose | | Descriptor: | AmyP, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | He, C, Liu, Y. | | Deposit date: | 2016-10-03 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a raw-starch-degrading bacterial alpha-amylase belonging to subfamily 37 of the glycoside hydrolase family GH13
Sci Rep, 7, 2017
|
|
5H05
 
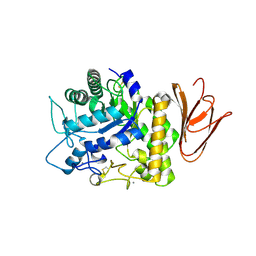 | | Crystal structure of AmyP E221Q in complex with MALTOTRIOSE | | Descriptor: | AmyP, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | He, C, Liu, Y. | | Deposit date: | 2016-10-03 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of a raw-starch-degrading bacterial alpha-amylase belonging to subfamily 37 of the glycoside hydrolase family GH13
Sci Rep, 7, 2017
|
|
2LA8
 
 | |
2MKV
 
 | |
8WBC
 
 | |
8WBB
 
 | |
8WBE
 
 | |
8WBD
 
 | |
8WBF
 
 | |
8WBG
 
 | |
8WBH
 
 | |
7FJC
 
 | | Crystal structure of SARS-CoV-2 Beta RBD complexed with P36-5D2 Fab | | Descriptor: | P36-5D2 heavy chain, P36-5D2 light chain, Spike protein S1, ... | | Authors: | Zhang, L.Q, Wang, X.Q, Shan, S.S, Lan, J. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | A Potent and Protective Human Neutralizing Antibody Against SARS-CoV-2 Variants.
Front Immunol, 12, 2021
|
|
7FAF
 
 | | S protein of SARS-CoV-2 in complex bound with P36-5D2 (state1) | | Descriptor: | P36-5D2 heavy chain, P36-5D2 light chain, Spike glycoprotein | | Authors: | Zhang, L, Wang, X, Zhang, S, Shan, S. | | Deposit date: | 2021-07-06 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | A Potent and Protective Human Neutralizing Antibody Against SARS-CoV-2 Variants.
Front Immunol, 12, 2021
|
|
7FAE
 
 | | S protein of SARS-CoV-2 in complex bound with P36-5D2(state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P36-5D2 heavy chain, ... | | Authors: | Zhang, L, Wang, X, Shan, S, Zhang, S. | | Deposit date: | 2021-07-06 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | A Potent and Protective Human Neutralizing Antibody Against SARS-CoV-2 Variants.
Front Immunol, 12, 2021
|
|
7XST
 
 | |
7XMX
 
 | | Cryo-EM structure of SARS-CoV-2 spike glycoprotein in complex with three F61 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F61 heavy chain, F61 light chain, ... | | Authors: | Wang, X, Li, X. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis of a two-antibody cocktail exhibiting highly potent and broadly neutralizing activities against SARS-CoV-2 variants including diverse Omicron sublineages.
Cell Discov, 8, 2022
|
|
7XMZ
 
 | | Cryo-EM structure of SARS-CoV-2 spike glycoprotein in complex with three D2 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D2 heavy chain, D2 light chain, ... | | Authors: | Wang, X, Li, X. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis of a two-antibody cocktail exhibiting highly potent and broadly neutralizing activities against SARS-CoV-2 variants including diverse Omicron sublineages.
Cell Discov, 8, 2022
|
|
