7DZK
 
 | | Fabp protein after hv | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZL
 
 | | A69C-M71L mutant of Fabp protein | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7YUB
 
 | | S1P-bound human SPNS2 | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, NbFab-H-chain, NbFab-L-chain, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
1KN5
 
 | | SOLUTION STRUCTURE OF ARID DOMAIN OF ADR6 FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | Transcription regulatory protein ADR6 | | Authors: | Tu, X, Wu, J, Xu, Y, Shi, Y. | | Deposit date: | 2001-12-18 | | Release date: | 2002-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments and secondary structure of ADR6 DNA-binding domain.
J.Biomol.Nmr, 21, 2001
|
|
1KKX
 
 | | Solution structure of the DNA-binding domain of ADR6 | | Descriptor: | Transcription regulatory protein ADR6 | | Authors: | Tu, X, Wu, J, Xu, Y, Shi, Y. | | Deposit date: | 2001-12-10 | | Release date: | 2002-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments and secondary structure of ADR6 DNA-binding domain.
J.Biomol.Nmr, 21, 2001
|
|
8CIJ
 
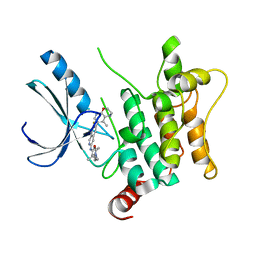 | | CRYSTAL STRUCTURE OF HUMAN HPK1 (MAP4K1) COMPLEX WITH 2-[8-Amino-7-fluoro-6-(8-methyl-2,3-dihydro-1H-pyrido[2,3-b][1,4]oxazin-7-yl)-isoquinolin-3-ylamino]-6-isopropyl-5,6-dihydro-4H-1,6,8a-triaza-azulen-7-one | | Descriptor: | 2-[[8-azanyl-7-fluoranyl-6-(8-methyl-2,3-dihydro-1~{H}-pyrido[2,3-b][1,4]oxazin-7-yl)isoquinolin-3-yl]amino]-6-propan-2-yl-5,8-dihydro-4~{H}-pyrazolo[1,5-d][1,4]diazepin-7-one, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Musil, D, Toure, M. | | Deposit date: | 2023-02-09 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.821 Å) | | Cite: | Discovery of quinazoline HPK1 inhibitors with high cellular potency.
Bioorg.Med.Chem., 92, 2023
|
|
8CDW
 
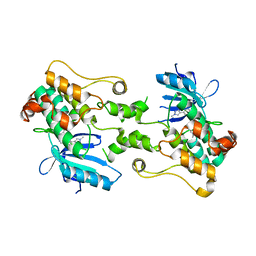 | |
6XBG
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW246 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-05 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XA4
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW241 | | Descriptor: | 3C-like proteinase, GLYCEROL, inhibitor UAW241 | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6K6R
 
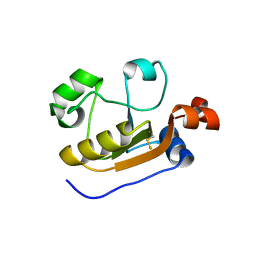 | |
7YUK
 
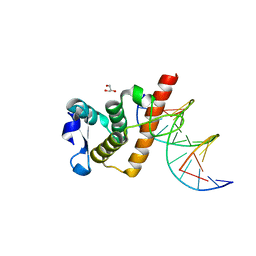 | | Complex structure of BANP BEN domain bound to DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*CP*GP*CP*GP*AP*GP*AP*G)-3'), GLYCEROL, Protein BANP | | Authors: | Zhang, J, Xiao, Y.Q, Chen, Y.X, Liu, K, Min, J.R. | | Deposit date: | 2022-08-17 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
7YUG
 
 | | Structure of human BANP BEN domain | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Zhang, J, Xiao, Y.Q, Chen, Y.X, Liu, K, Min, J.R. | | Deposit date: | 2022-08-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
7YUL
 
 | | Crystal structure of human BEND6 BEN domain in complex with DNA | | Descriptor: | BEN domain-containing protein 6, DNA (5'-D(*CP*TP*CP*TP*CP*GP*CP*GP*AP*GP*AP*G)-3'), GLYCOLIC ACID | | Authors: | Liu, K, Xiao, Y.Q, Zhang, J, Min, J.R. | | Deposit date: | 2022-08-17 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
7YUN
 
 | | Crystal structure of human BEND6 BEN domain in complex with methylated DNA | | Descriptor: | BEN domain-containing protein 6, DNA (5'-D(*CP*TP*CP*TP*CP*GP*(5CM)P*GP*AP*GP*AP*G)-3') | | Authors: | Liu, K, Xiao, Y.Q, Zhang, J, Min, J.R. | | Deposit date: | 2022-08-17 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
7YFS
 
 | | The NMR structure of noursin, a tricyclic ribosomal peptide containing a histidine-to-butyrine crosslink | | Descriptor: | noursin | | Authors: | Yao, H, Li, Y, Zhang, T, Gao, J, Wang, H. | | Deposit date: | 2022-07-09 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Discovery and biosynthesis of tricyclic copper-binding ribosomal peptides containing histidine-to-butyrine crosslinks.
Nat Commun, 14, 2023
|
|
7VKT
 
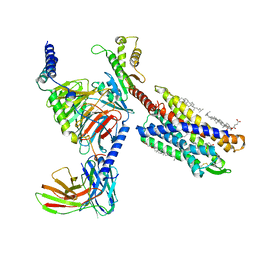 | | cryo-EM structure of LTB4-bound BLT1 in complex with Gi protein | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Wang, N. | | Deposit date: | 2021-10-01 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of leukotriene B4 receptor 1 activation.
Nat Commun, 13, 2022
|
|
7W8L
 
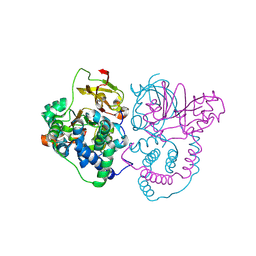 | | Crystal Structure of Co-type nitrile hydratase mutant from Pseudonocardia thermophila - M46R | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit beta, Nitrile hydratase | | Authors: | Ma, D, Cheng, Z.Y, Hou, X.D, Peplowski, L, Lai, Q.P, Fu, K, Yin, D.J, Rao, Y.J, Zhou, Z.M. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Insight into the broadened substrate scope of nitrile hydratase by static and dynamic structure analysis.
Chem Sci, 13, 2022
|
|
7W8M
 
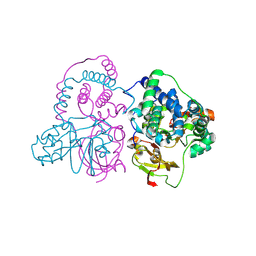 | | Crystal structure of Co-type nitrile hydratase mutant from Pseudomonas thermophila - A129R | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit beta, Nitrile hydratase | | Authors: | Ma, D, Cheng, Z.Y, Hou, X.D, Peplowski, L, Lai, Q.P, Fu, K, Yin, D.J, Rao, Y.J, Zhou, Z.M. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the broadened substrate scope of nitrile hydratase by static and dynamic structure analysis.
Chem Sci, 13, 2022
|
|
7N9C
 
 | |
7N9E
 
 | |
7N9B
 
 | |
7N9T
 
 | |
6OAQ
 
 | | Crystal structure of a dual sensor histidine kinase in BeF3- bound state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Dual sensor histidine kinase, MAGNESIUM ION, ... | | Authors: | Heewhan, S, Zhong, R, Xiaoli, Z, Sepalika, B, Xiaojing, Y. | | Deposit date: | 2019-03-18 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural basis of molecular logic OR in a dual-sensor histidine kinase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OB8
 
 | | Crystal structure of a dual sensor histidine kinase in green light illuminated state | | Descriptor: | Dual sensor histidine kinase, MAGNESIUM ION, PHYCOCYANOBILIN | | Authors: | Heewhan, S, Zhong, R, Xiaoli, Z, Sepalika, B, Xiaojing, Y. | | Deposit date: | 2019-03-19 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of molecular logic OR in a dual-sensor histidine kinase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7YEU
 
 | | Superfolder green fluorescent protein with phosphine unnatural amino acid P3BF | | Descriptor: | Superfolder green fluorescent protein | | Authors: | Hu, C, Duan, H.Z, Liu, X.H, Chen, Y.X, Wang, J.Y. | | Deposit date: | 2022-07-06 | | Release date: | 2023-07-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Genetically Encoded Phosphine Ligand for Metalloprotein Design.
J.Am.Chem.Soc., 144, 2022
|
|
