6MPH
 
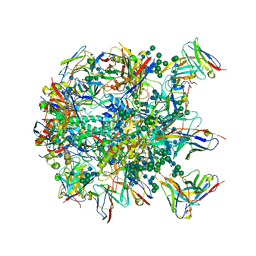 | | Cryo-EM structure at 3.8 A resolution of HIV-1 fusion peptide-directed antibody, DF1W-a.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DF1W-a.01 Light chain, ... | | Authors: | Acharya, P, Xu, K, Kwong, P.D. | | Deposit date: | 2018-10-06 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
6MQM
 
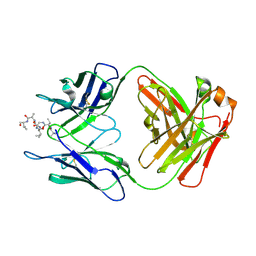 | |
6N16
 
 | |
6N1W
 
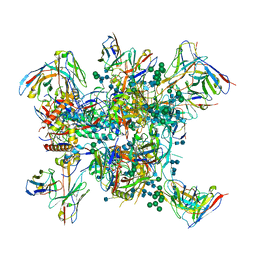 | |
6NF2
 
 | |
6MQE
 
 | |
6MQS
 
 | |
6MQR
 
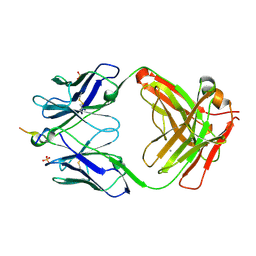 | | Vaccine-elicited NHP FP-targeting neutralizing antibody 0PV-a.01 in complex with FP (residue 512-519) | | Descriptor: | CALCIUM ION, HIV fusion peptide residue 512-519, SULFATE ION, ... | | Authors: | Xu, K, Wang, Y, Kwong, P.D. | | Deposit date: | 2018-10-10 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
6N1V
 
 | |
6MPG
 
 | | Cryo-EM structure at 3.2 A resolution of HIV-1 fusion peptide-directed antibody, A12V163-b.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A12V163-b.01 Heavy Chain, ... | | Authors: | Acharya, P, Kwong, P.D. | | Deposit date: | 2018-10-06 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
5WXB
 
 | |
3DCX
 
 | |
3DUE
 
 | |
4ME2
 
 | | Crystal Structure of THA8 protein from Brachypodium distachyon | | Descriptor: | Uncharacterized protein | | Authors: | Ke, J, Chen, R.Z, Ban, T, Brunzelle, J.S, Gu, X, Melcher, K, Xu, H.E. | | Deposit date: | 2013-08-24 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for RNA recognition by a dimeric PPR-protein complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
5NXY
 
 | | Crystal structure of OpuAC from B. subtilis in complex with Arsenobetaine | | Descriptor: | 1,2-ETHANEDIOL, 2-(trimethyl-lambda~5~-arsanyl)ethanol, Osmotically activated L-carnitine/choline ABC transporter substrate-binding protein OpuCC | | Authors: | Hofmann, T, Bremer, E, Schmitt, L, Smits, S. | | Deposit date: | 2017-05-11 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Arsenobetaine: an ecophysiologically important organoarsenical confers cytoprotection against osmotic stress and growth temperature extremes.
Environ. Microbiol., 20, 2018
|
|
5NXX
 
 | | Crystal structure of OpuAC from B. subtilis in complex with Arsenobetaine | | Descriptor: | (trimethylarsonio)acetate, Glycine betaine ABC transport system glycine betaine-binding protein OpuAC | | Authors: | Hofmann, T, Bremer, E, Schmitt, L, Smits, S. | | Deposit date: | 2017-05-11 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Arsenobetaine: an ecophysiologically important organoarsenical confers cytoprotection against osmotic stress and growth temperature extremes.
Environ. Microbiol., 20, 2018
|
|
4N2Q
 
 | | Crystal structure of THA8 in complex with Zm4 RNA | | Descriptor: | RNA (5'-R(*AP*AP*GP*AP*AP*GP*AP*AP*AP*UP*UP*GP*G)-3'), THA8 RNA binding protein | | Authors: | Ke, J, Chen, R.Z, Ban, T, Zhou, X.E, Gu, X, Brunzelle, J.S, Zhu, J.K, Melcher, K, Xu, H.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for RNA recognition by a dimeric PPR-protein complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4N2S
 
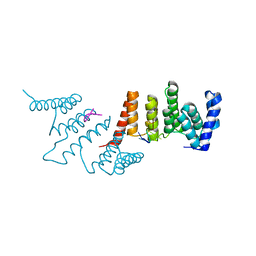 | | Crystal Structure of THA8 in complex with Zm1a-6 RNA | | Descriptor: | THA8 RNA binding protein, Zm1a-6 RNA | | Authors: | Ke, J, Chen, R.Z, Ban, T, Zhou, X.E, Gu, X, Brunzelle, J.S, Zhu, J.K, Melcher, K, Xu, H.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for RNA recognition by a dimeric PPR-protein complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
8EE7
 
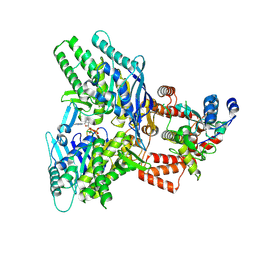 | |
8EE4
 
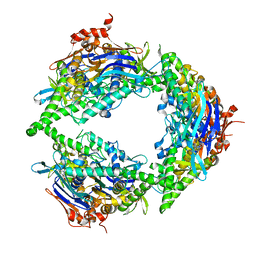 | | Structure of PtuA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PtuA | | Authors: | Shen, Z.F, Fu, T.M. | | Deposit date: | 2022-09-06 | | Release date: | 2024-01-03 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | PtuA and PtuB assemble into an inflammasome-like oligomer for anti-phage defense.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8EEA
 
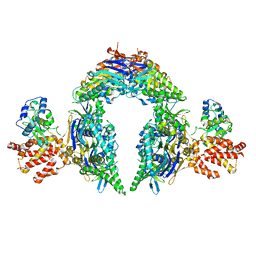 | | Structure of E.coli Septu (PtuAB) complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PtuA, PtuB | | Authors: | Shen, Z.F, Fu, T.M. | | Deposit date: | 2022-09-06 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | PtuA and PtuB assemble into an inflammasome-like oligomer for anti-phage defense.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5DMR
 
 | |
5DMQ
 
 | |
8Y58
 
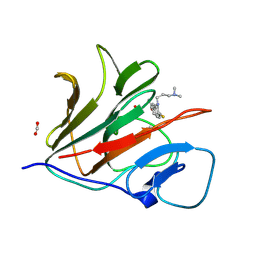 | | Crystal structure of TRIM21 PRYSPRY (D355A) in complex with acepromazine. | | Descriptor: | 1-[10-(3-DIMETHYLAMINO-PROPYL)-10H-PHENOTHIAZIN-2-YL]-ETHANONE, E3 ubiquitin-protein ligase TRIM21, FORMIC ACID | | Authors: | Lu, P, Cheng, Y, Xue, L, Ren, X, Huang, N, Han, T. | | Deposit date: | 2024-01-31 | | Release date: | 2024-10-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective degradation of multimeric proteins by TRIM21-based molecular glue and PROTAC degraders.
Cell, 187, 2024
|
|
8Y59
 
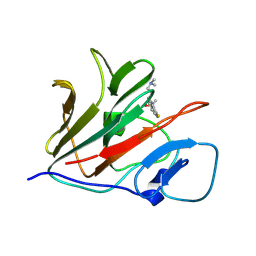 | | Crystal structure of TRIM21 PRYSPRY (D355A) in complex with (S)-hydroxyl-acepromazine. | | Descriptor: | (1~{S})-1-[10-[3-(dimethylamino)propyl]phenothiazin-2-yl]ethanol, E3 ubiquitin-protein ligase TRIM21 | | Authors: | Lu, P, Cheng, Y, Xue, L, Ren, X, Huang, N, Han, T. | | Deposit date: | 2024-01-31 | | Release date: | 2024-10-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Selective degradation of multimeric proteins by TRIM21-based molecular glue and PROTAC degraders.
Cell, 187, 2024
|
|
