6YEL
 
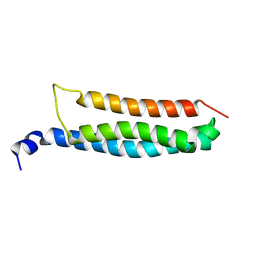 | | Stromal interaction molecule 1 coiled-coil 1 fragment | | Descriptor: | Stromal interaction molecule 1 | | Authors: | Rathner, P, Cerofolini, L, Ravera, E, Bechmann, M, Grabmayr, H, Fahrner, M, Fragai, M, Romanin, C, Luchinat, C, Mueller, N. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-02 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Interhelical interactions within the STIM1 CC1 domain modulate CRAC channel activation.
Nat.Chem.Biol., 17, 2021
|
|
8RE0
 
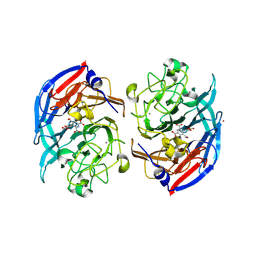 | | Soluble glucose dehydrogenase from acinetobacter calcoaceticus - double mutant pH8 | | Descriptor: | 3-(3,5-dicarboxy-1~{H}-pyrrol-2-yl)pyridine-2,4,6-tricarboxylic acid, CALCIUM ION, LITHIUM ION, ... | | Authors: | Lublin, V, Chavas, L, Stines-Chaumeil, C, Kauffmann, B, Giraud, M.F, Thompson, A. | | Deposit date: | 2023-12-09 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Does Acinetobacter calcoaceticus glucose dehydrogenase produce self-damaging H2O2?
Biosci.Rep., 44, 2024
|
|
8RFK
 
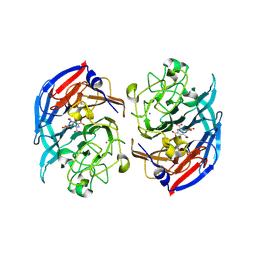 | | Soluble glucose dehydrogenase from acinetobacter calcoaceticus - single mutant pH8 | | Descriptor: | 3-(3,5-dicarboxy-1~{H}-pyrrol-2-yl)pyridine-2,4,6-tricarboxylic acid, CALCIUM ION, Quinoprotein glucose dehydrogenase B | | Authors: | Lublin, V, Chavas, L, Stines-Chaumeil, C, Kauffmann, B, Giraud, M.F, Thompson, A. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Does Acinetobacter calcoaceticus glucose dehydrogenase produce self-damaging H2O2?
Biosci.Rep., 44, 2024
|
|
8HSY
 
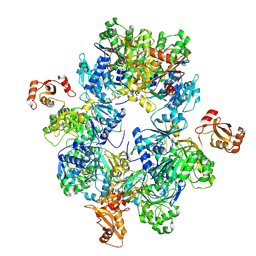 | | Acyl-ACP Synthetase structure | | Descriptor: | Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2022-12-20 | | Release date: | 2023-12-27 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | An inhibitory mechanism of AasS, an exogenous fatty acid scavenger: Implications for re-sensitization of FAS II antimicrobials.
Plos Pathog., 20, 2024
|
|
8RG1
 
 | | Soluble glucose dehydrogenase from acinetobacter calcoaceticus - wild type pH8 | | Descriptor: | 3-(3,5-dicarboxy-1~{H}-pyrrol-2-yl)pyridine-2,4,6-tricarboxylic acid, CALCIUM ION, LITHIUM ION, ... | | Authors: | Lublin, V, Chavas, L, Stines-Chaumeil, C, Kauffmann, B, Giraud, M.F, Thompson, A. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Does Acinetobacter calcoaceticus glucose dehydrogenase produce self-damaging H2O2?
Biosci.Rep., 44, 2024
|
|
8REV
 
 | | Structure of XPD stalled at a Y-fork DNA containing a interstrand crosslink | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase CHL1, DNA (46-MER), ... | | Authors: | Kuper, J, Hove, T, Kisker, C. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | XPD stalled on cross-linked DNA provides insight into damage verification.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8S4J
 
 | | Structure, substrate selectivity determinants and membrane interactions of a Glutamate-specific TAXI TRAP binding protein from Vibrio cholerae. | | Descriptor: | GLUTAMIC ACID, SODIUM ION, TAXI family TRAP transporter solute-binding subunit | | Authors: | Mulligan, C, Daab, A, Davies, J. | | Deposit date: | 2024-02-21 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and selectivity of a glutamate-specific TAXI TRAP binding protein from Vibrio cholerae.
J.Gen.Physiol., 156, 2024
|
|
3CX5
 
 | |
8RRY
 
 | | Crystal structure of copper-loaded SmAA10 | | Descriptor: | CBP21, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Munzone, A, Pujol, M, Reglier, M, Royant, A, Simaan, A.J, Decroos, C. | | Deposit date: | 2024-01-24 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45561326 Å) | | Cite: | Integrated Experimental and Theoretical Investigation of Copper Active Site Properties of a Lytic Polysaccharide Monooxygenase from Serratia marcescens.
Inorg.Chem., 63, 2024
|
|
3DB4
 
 | | Crystal structure of the tandem tudor domains of the E3 ubiquitin-protein ligase UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, SULFATE ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Dong, A, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-30 | | Release date: | 2008-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein.
J.Biol.Chem., 286, 2011
|
|
6ZB5
 
 | | SARS CoV-2 Spike protein, Closed conformation, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Burucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2020-06-07 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein.
Science, 370, 2020
|
|
3DJS
 
 | |
3DJZ
 
 | |
6ZFC
 
 | | Fucose-binding lectin from Burkholderia ambifaria (BamBL) in complex with a fucosyl derivative | | Descriptor: | 2-[(2~{S},3~{S},4~{R},4~{a}~{S},10~{a}~{S})-2-methyl-3,4-bis(oxidanyl)-3,4,4~{a},10~{a}-tetrahydro-2~{H}-pyrano[2,3-b][1,4]benzoxathiin-7-yl]-~{N}-(3-oxidanylpropyl)ethanamide, bacterial lectin from Burkholderia ambifaria | | Authors: | Kuhaudomlarp, S, Gillon, E, Fragai, M, Cerofolini, L, Giuntini, S, Denis, M, Santarsia, S, Valori, C, Dondoni, A, Fallarini, S, Lombardi, G, Nativi, C, Imberty, A. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Fucosylated ubiquitin and orthogonally glycosylated mutant A28C: conceptually new ligands for Burkholderia ambifaria lectin (BambL).
Chem Sci, 11, 2020
|
|
4UDB
 
 | | MR in complex with desisobutyrylciclesonide | | Descriptor: | DESISOBUYTYRYL CICLESONIDE, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, JellesmarkJensen, T, Cavallin, A, Nilsson, E, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
6ZB4
 
 | | SARS CoV-2 Spike protein, Closed conformation, C1 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Burucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein.
Science, 370, 2020
|
|
8RO1
 
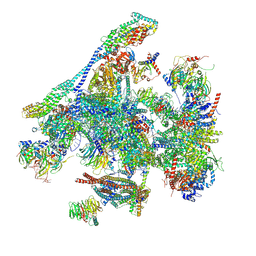 | | Structure of the C. elegans Intron Lariat Spliceosome double-primed for disassembly (ILS'') | | Descriptor: | CWF19-like protein 1 homolog, CWF19-like protein 2 homolog, Cell division cycle 5-like protein, ... | | Authors: | Vorlaender, M.K, Rothe, P, Plaschka, C. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism for the initiation of spliceosome disassembly.
Nature, 632, 2024
|
|
8B5G
 
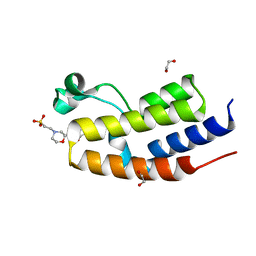 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 7,8-dimethoxy-3-methyl-1,3-dihydro-2H-benzo[d]azepin-2-one | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7,8-dimethoxy-3-methyl-1~{H}-3-benzazepin-2-one, ... | | Authors: | Chung, C. | | Deposit date: | 2022-09-22 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.619 Å) | | Cite: | Identification and Optimization of a Ligand-Efficient Benzoazepinone Bromodomain and Extra Terminal (BET) Family Acetyl-Lysine Mimetic into the Oral Candidate Quality Molecule I-BET432.
J.Med.Chem., 65, 2022
|
|
8B5H
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH (R)-7-((R)-1,2-dihydroxyethyl)-1,3-dimethyl-5-(1-methyl-1H-pyrazol-4-yl)-1,3-dihydro-2H-benzo[d]azepin-2-one | | Descriptor: | (1~{R})-7-[(1~{R})-1,2-bis(oxidanyl)ethyl]-1,3-dimethyl-5-(1-methylpyrazol-4-yl)-1~{H}-3-benzazepin-2-one, 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, ... | | Authors: | Chung, C. | | Deposit date: | 2022-09-22 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Identification and Optimization of a Ligand-Efficient Benzoazepinone Bromodomain and Extra Terminal (BET) Family Acetyl-Lysine Mimetic into the Oral Candidate Quality Molecule I-BET432.
J.Med.Chem., 65, 2022
|
|
8B5I
 
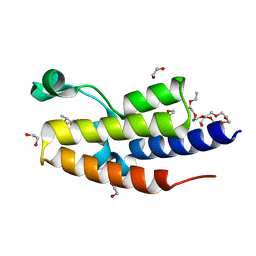 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 7,8-dimethoxy-1,3-dimethyl-1,3-dihydro-2H-benzo[d]azepin-2-one | | Descriptor: | (1~{R})-7,8-dimethoxy-1,3-dimethyl-1~{H}-3-benzazepin-2-one, 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Chung, C. | | Deposit date: | 2022-09-22 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Identification and Optimization of a Ligand-Efficient Benzoazepinone Bromodomain and Extra Terminal (BET) Family Acetyl-Lysine Mimetic into the Oral Candidate Quality Molecule I-BET432.
J.Med.Chem., 65, 2022
|
|
8B5J
 
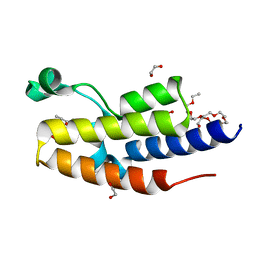 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 7,8-dimethoxy-1,3-dimethyl-1,3-dihydro-2H-benzo[d]azepin-2-one | | Descriptor: | (5~{R})-7,8-dimethoxy-3,5-dimethyl-2,5-dihydro-1~{H}-3-benzazepin-4-one, 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Chung, C. | | Deposit date: | 2022-09-22 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Identification and Optimization of a Ligand-Efficient Benzoazepinone Bromodomain and Extra Terminal (BET) Family Acetyl-Lysine Mimetic into the Oral Candidate Quality Molecule I-BET432.
J.Med.Chem., 65, 2022
|
|
5Y6N
 
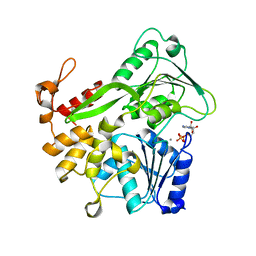 | | Zika virus helicase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Helicase domain from Genome polyprotein, MANGANESE (II) ION | | Authors: | Yang, X.Y, Chen, C, Tian, H.L, Chi, H, Mu, Z.Y, Zhang, T.Q, Yang, K.L, Zhao, Q, Liu, X.H, Wang, Z.F, Ji, X.Y, Yang, H.T. | | Deposit date: | 2017-08-12 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Mechanism of ATP hydrolysis by the Zika virus helicase.
FASEB J., 32, 2018
|
|
4V4P
 
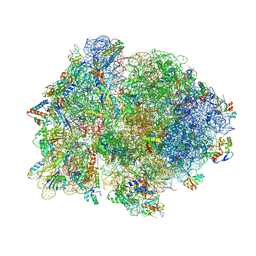 | | Crystal structure of 70S ribosome with thrS operator and tRNAs. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Jenner, L, Romby, P, Rees, B, Schulze-Briese, C, Springer, M, Ehresmann, C, Ehresmann, B, Moras, D, Yusupova, G, Yusupov, M. | | Deposit date: | 2005-01-19 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Translational operator of mRNA on the ribosome: how repressor proteins exclude ribosome binding.
Science, 308, 2005
|
|
4UV4
 
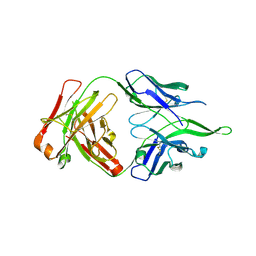 | | Crystal structure of anti-FPR Fpro0165 Fab fragment | | Descriptor: | FPRO0165 FAB | | Authors: | Douthwaite, J.A, Sridharan, S, Huntington, C, Marwood, R, Hammersley, J, Hakulinen, J.K, Ek, M, Sjogren, T, Rider, D, Privezentzev, C, Seaman, J.C, Cariuk, P, Knights, V, Young, J, Wilkinson, T, Sleeman, M, Finch, D.K, Lowe, D.C, Vaughan, T.J. | | Deposit date: | 2014-08-04 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Affinity Maturation of a Novel Antagonistic Human Monoclonal Antibody with a Long Vh Cdr3 Targeting the Class a Gpcr Formyl-Peptide Receptor 1.
Mabs, 7, 2015
|
|
3JXE
 
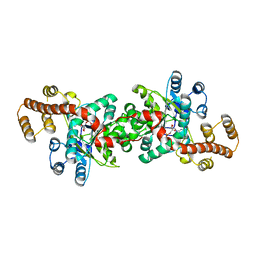 | | Crystal structure of Pyrococcus horikoshii tryptophanyl-tRNA synthetase in complex with TrpAMP | | Descriptor: | SULFATE ION, TRYPTOPHANYL-5'AMP, Tryptophanyl-tRNA synthetase | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Yang, B, Ding, J. | | Deposit date: | 2009-09-19 | | Release date: | 2009-11-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of P. horikoshii tryptophanyl-tRNA synthetase and structure-based phylogenetic analysis suggest an archaeal origin of tryptophanyl-tRNA synthetase
To be Published
|
|
