8I1G
 
 | |
8I8T
 
 | |
8I19
 
 | |
6K9U
 
 | | Discovery of Pyrazolo[1,5-a]pyrimidine Derivative as a Highly Selective PDE10A Inhibitor | | Descriptor: | 2-(3,7-dimethylquinoxalin-2-yl)-~{N}-(oxan-4-yl)-5-pyrrolidin-1-yl-pyrazolo[1,5-a]pyrimidin-7-amine, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Takedomi, K, Koizumi, Y. | | Deposit date: | 2019-06-18 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of a pyrazolo[1,5-a]pyrimidine derivative (MT-3014) as a highly selective PDE10A inhibitor via core structure transformation from the stilbene moiety.
Bioorg.Med.Chem., 27, 2019
|
|
6JTC
 
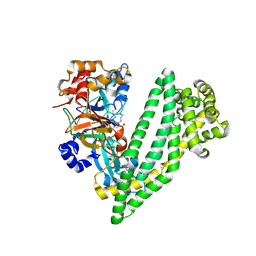 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) with SH-5 from Porphyromonas gingivalis (Space) | | Descriptor: | 2-(2-azanylethylamino)-5-nitro-benzoic acid, Asp/Glu-specific dipeptidyl-peptidase, GLYCEROL | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Roppongi, S, Kushibiki, C, Nakamura, A, Ogasawara, W, Tanaka, N. | | Deposit date: | 2019-04-10 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Fragment-based discovery of the first nonpeptidyl inhibitor of an S46 family peptidase.
Sci Rep, 9, 2019
|
|
6FD7
 
 | |
6FDT
 
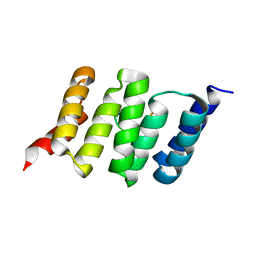 | |
5X2M
 
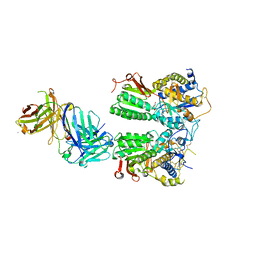 | | Crystal structure of the medaka fish taste receptor T1r2a-T1r3 ligand binding domains in complex with L-glutamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Nuemket, N, Yasui, N, Atsumi, N, Yamashita, A. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structural basis for perception of diverse chemical substances by T1r taste receptors
Nat Commun, 8, 2017
|
|
3ONE
 
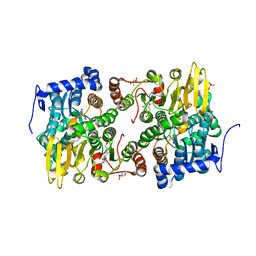 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with adenine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6GXZ
 
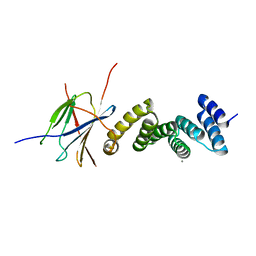 | | Crystal structure of the human RPAP3(TPR2)-PIH1D1(CS) complex | | Descriptor: | CALCIUM ION, PIH1 domain-containing protein 1, RNA polymerase II-associated protein 3 | | Authors: | Henri, J, Quinternet, M, Manival, X, Chagot, M.-E, Charpentier, B, Meyer, P. | | Deposit date: | 2018-06-27 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.965 Å) | | Cite: | Deep Structural Analysis of RPAP3 and PIH1D1, Two Components of the HSP90 Co-chaperone R2TP Complex.
Structure, 26, 2018
|
|
5X2N
 
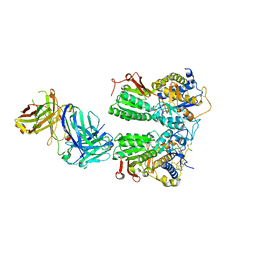 | | Crystal structure of the medaka fish taste receptor T1r2a-T1r3 ligand binding domains in complex with L-alanine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALANINE, CHLORIDE ION, ... | | Authors: | Nuemket, N, Yasui, N, Atsumi, N, Yamashita, A. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for perception of diverse chemical substances by T1r taste receptors
Nat Commun, 8, 2017
|
|
5X2P
 
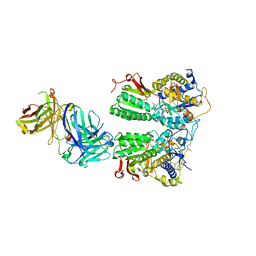 | | Crystal structure of the medaka fish taste receptor T1r2a-T1r3 ligand binding domains in complex with L-glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Nuemket, N, Yasui, N, Atsumi, N, Yamashita, A. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | Structural basis for perception of diverse chemical substances by T1r taste receptors
Nat Commun, 8, 2017
|
|
5X2Q
 
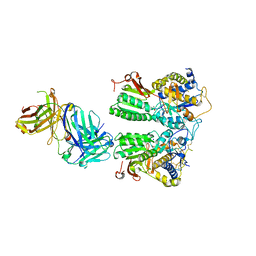 | | Crystal structure of the medaka fish taste receptor T1r2a-T1r3 ligand binding domains in complex with glycine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Nuemket, N, Yasui, N, Atsumi, N, Yamashita, A. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for perception of diverse chemical substances by T1r taste receptors
Nat Commun, 8, 2017
|
|
5X2O
 
 | | Crystal structure of the medaka fish taste receptor T1r2a-T1r3 ligand binding domains in complex with L-arginine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ARGININE, CALCIUM ION, ... | | Authors: | Nuemket, N, Yasui, N, Atsumi, N, Yamashita, A. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for perception of diverse chemical substances by T1r taste receptors
Nat Commun, 8, 2017
|
|
3OND
 
 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with adenosine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3ONF
 
 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with cordycepin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3'-DEOXYADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6LX6
 
 | | X-ray structure of human PPARalpha ligand binding domain-palmitic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | GLYCEROL, PALMITIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LXB
 
 | | X-ray structure of human PPARalpha ligand binding domain-saroglitazar co-crystals obtained by soaking | | Descriptor: | (2S)-2-ethoxy-3-[4-[2-[2-methyl-5-(4-methylsulfanylphenyl)pyrrol-1-yl]ethoxy]phenyl]propanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Honda, A, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LXA
 
 | | X-ray structure of human PPARalpha ligand binding domain-eicosapentaenoic acid (EPA) co-crystals obtained by delipidation and cross-seeding | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
5E6O
 
 | | Crystal structure of C. elegans LGG-2 bound to an AIM/LIR motif | | Descriptor: | Protein lgg-2, TRP-GLU-GLU-LEU | | Authors: | Qi, X, Ren, J.Q, Wu, F, Zhang, H, Feng, W. | | Deposit date: | 2015-10-10 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy
Mol.Cell, 60, 2015
|
|
6LX7
 
 | | X-ray structure of human PPARalpha ligand binding domain-stearic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | GLYCEROL, Peroxisome proliferator-activated receptor alpha, STEARIC ACID | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LXC
 
 | | X-ray structure of human PPARalpha ligand binding domain-saroglitazar co-crystals obtained by delipidation and cross-seeding | | Descriptor: | (2S)-2-ethoxy-3-[4-[2-[2-methyl-5-(4-methylsulfanylphenyl)pyrrol-1-yl]ethoxy]phenyl]propanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Honda, A, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
5E6N
 
 | | Crystal structure of C. elegans LGG-2 | | Descriptor: | Protein lgg-2 | | Authors: | Qi, X, Ren, J.Q, Wu, F, Zhang, H, Feng, W. | | Deposit date: | 2015-10-10 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy
Mol.Cell, 60, 2015
|
|
2ZFA
 
 | | Structure of Lactate Oxidase at pH4.5 from AEROCOCCUS VIRIDANS | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Lactate oxidase | | Authors: | Furuichi, M, Balasundaresan, D, Suzuki, N, Yoshida, Y, Minagawa, H, Kaneko, H, Waga, I, Kumar, P.K.R, Mizuno, H. | | Deposit date: | 2007-12-26 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray structures of Aerococcus viridans lactate oxidase and its complex with D-lactate at pH 4.5 show an alpha-hydroxyacid oxidation mechanism
J.Mol.Biol., 378, 2008
|
|
7X91
 
 | |
