7WU4
 
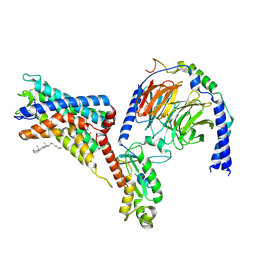 | | Cryo-EM structure of the adhesion GPCR ADGRF1 in complex with miniGi | | Descriptor: | Adhesion G-protein coupled receptor F1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-05 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7WU2
 
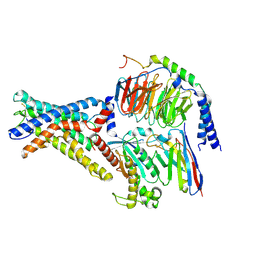 | | Cryo-EM structure of the adhesion GPCR ADGRD1 in complex with miniGs | | Descriptor: | Adhesion G-protein coupled receptor D1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-05 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7WU5
 
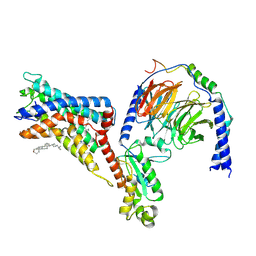 | | Cryo-EM structure of the adhesion GPCR ADGRF1(H565A/T567A) in complex with miniGi | | Descriptor: | Adhesion G-protein coupled receptor F1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-05 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7WU3
 
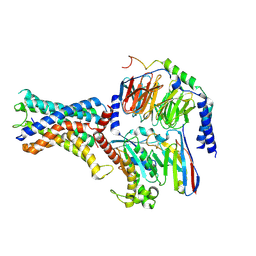 | | Cryo-EM structure of the adhesion GPCR ADGRF1 in complex with miniGs | | Descriptor: | Adhesion G-protein coupled receptor F1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-05 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7D7L
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in complex with YM155 | | Descriptor: | 1-(2-methoxyethyl)-2-methyl-3-(pyrazin-2-ylmethyl)benzo[f]benzimidazol-3-ium-4,9-dione, CAFFEINE, GLYCEROL, ... | | Authors: | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2020-10-04 | | Release date: | 2021-04-21 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
7D7K
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in apo form | | Descriptor: | 1,2-ETHANEDIOL, CAFFEINE, Non-structural protein 3, ... | | Authors: | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2020-10-04 | | Release date: | 2021-04-21 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
7DPM
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW06 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-12-20 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.304 Å) | | Cite: | Characterization of MW06, a human monoclonal antibody with cross-neutralization activity against both SARS-CoV-2 and SARS-CoV.
Mabs, 13, 2021
|
|
2P4U
 
 | | Crystal structure of acid phosphatase 1 (Acp1) from Mus musculus | | Descriptor: | Acid phosphatase 1, PHOSPHATE ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Wu, B, Xu, W, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-13 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
6DRU
 
 | | Xylosidase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Glycosyl hydrolases family 31 family protein, ... | | Authors: | Cao, H, Xu, W, Betancourt, M, Walton, J.D, Brumm, P, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of alpha-Xylosidase fromAspergillus nigerin Complex with a Hydrolyzed Xyloglucan Product and New Insights in Accurately Predicting Substrate Specificities of GH31 Family Glycosidases.
Acs Sustain Chem Eng, 8, 2020
|
|
2NV5
 
 | | Crystal structure of a C-terminal phosphatase domain of Rattus norvegicus ortholog of human protein tyrosine phosphatase, receptor type, D (PTPRD) | | Descriptor: | PTPRD, PHOSPHATASE | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Iizuka, M, Xu, W, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-10 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2P8E
 
 | | Crystal structure of the serine/threonine phosphatase domain of human PPM1B | | Descriptor: | MAGNESIUM ION, PPM1B beta isoform variant 6 | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Lau, C, Xu, W, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-22 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
3T4F
 
 | | Crystal Structure of a KGE Collagen Mimetic Peptide at 1.68 A | | Descriptor: | collagen mimetic peptide | | Authors: | Fallas, J.A, Dong, J, Miller, M.D, Tao, Y.J, Hartgerink, J.D. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural insights into charge pair interactions in triple helical collagen-like proteins.
J.Biol.Chem., 287, 2012
|
|
3U29
 
 | | Crystal Structure of a KGD Collagen Mimetic Peptide at 2.0 A | | Descriptor: | collagen mimetic peptide | | Authors: | Fallas, J.A, Dong, J, Miller, M.D, Tao, Y.J, Hartgerink, J.D. | | Deposit date: | 2011-10-02 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into charge pair interactions in triple helical collagen-like proteins.
J.Biol.Chem., 287, 2012
|
|
7VXH
 
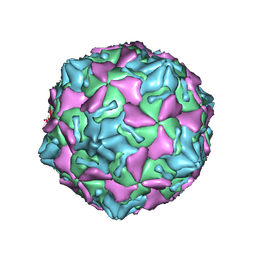 | | Coxsackievirus B3 full particle at pH7.4 (VP3-234Q) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Wang, Q.L, Liu, C.C. | | Deposit date: | 2021-11-12 | | Release date: | 2022-01-19 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VY0
 
 | | Coxsackievirus B3 full particle at pH7.4 (VP3-234N) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Wang, Q.L, Liu, C.C. | | Deposit date: | 2021-11-13 | | Release date: | 2022-01-19 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VYM
 
 | |
7VY5
 
 | | Coxsackievirus B3 (VP3-234Q) incubation with CD55 at pH7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Wang, Q.L, Liu, C.C. | | Deposit date: | 2021-11-13 | | Release date: | 2022-01-19 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VY6
 
 | | Coxsackievirus B3(VP3-234N) incubate with CD55 at pH7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Wang, Q.L, Liu, C.C. | | Deposit date: | 2021-11-13 | | Release date: | 2022-01-19 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
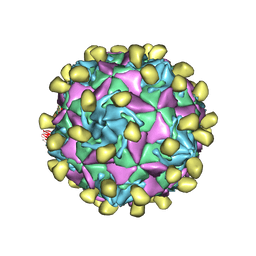
7VXZ
 
 | |
7VYL
 
 | |
7VYK
 
 | |
7C2M
 
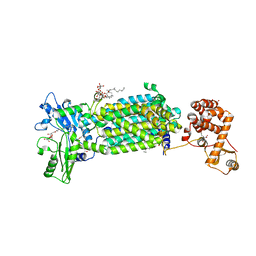 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with NITD-349 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Chimera of drug exporters of the RND superfamily-like protein and Endolysin, N-(4,4-dimethylcyclohexyl)-4,6-bis(fluoranyl)-1H-indole-2-carboxamide, ... | | Authors: | Zhang, B, Yang, X, Hu, T, Rao, Z. | | Deposit date: | 2020-05-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for the Inhibition of Mycobacterial MmpL3 by NITD-349 and SPIRO.
J.Mol.Biol., 432, 2020
|
|
7CAE
 
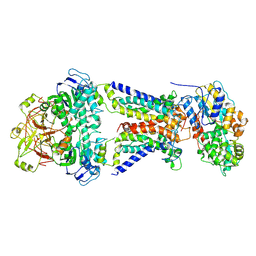 | | Mycobacterium smegmatis LpqY-SugABC complex in the resting state | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liu, F, Liang, J, Zhang, B, Gao, Y, Yang, X, Hu, T, Rao, Z. | | Deposit date: | 2020-06-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural basis of trehalose recycling by the ABC transporter LpqY-SugABC.
Sci Adv, 6, 2020
|
|
7C53
 
 | |
7W17
 
 | | Coxsackievirus B3 full particle at pH7.4 (VP3-234E) | | Descriptor: | PALMITIC ACID, VP1, VP2, ... | | Authors: | Wang, Q.L, Liu, C.C. | | Deposit date: | 2021-11-19 | | Release date: | 2022-01-19 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of differential receptor usage for naturally occurring CD55-binding and -nonbinding coxsackievirus B3 strains.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
