4RQQ
 
 | |
1WKO
 
 | |
1WKP
 
 | | Flowering locus t (ft) from arabidopsis thaliana | | Descriptor: | FLOWERING LOCUS T protein, SULFATE ION | | Authors: | Miller, D, Banfield, M.J, Winter, V.J, Brady, R.L. | | Deposit date: | 2004-06-01 | | Release date: | 2005-06-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A divergent external loop confers antagonistic activity on floral regulators FT and TFL1.
Embo J., 25, 2006
|
|
4B18
 
 | |
4ES0
 
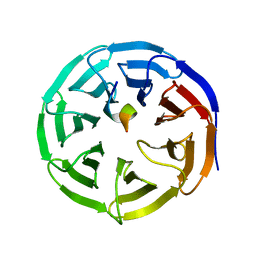 | | X-ray structure of WDR5-SETd1b Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase SETD1B, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-21 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.817 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4ERZ
 
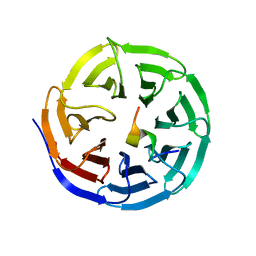 | | X-ray structure of WDR5-MLL4 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL4, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-21 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4EWR
 
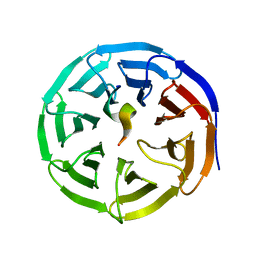 | | X-ray structure of WDR5-SETd1a Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase SETD1A, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-27 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4ESG
 
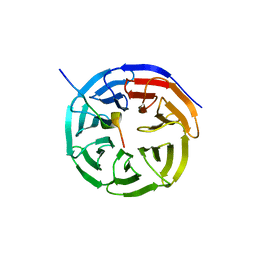 | | X-ray structure of WDR5-MLL1 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4ERY
 
 | | X-ray structure of WDR5-MLL3 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL3, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-21 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
4ERQ
 
 | | X-ray structure of WDR5-MLL2 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL2, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
7YPC
 
 | | Notothenia coriiceps TRAF4 | | Descriptor: | TNF receptor-associated factor | | Authors: | Park, H.H, Kim, C.M, Jang, H.S. | | Deposit date: | 2022-08-03 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structure of fish TRAF4 and its implication in TRAF4-mediated immune cell and platelet signaling.
Fish Shellfish Immunol., 132, 2023
|
|
5JCI
 
 | | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Os09g0567300 protein | | Authors: | Park, A.K, Kim, H.W. | | Deposit date: | 2016-04-15 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica
Sci Rep, 6, 2016
|
|
5JCM
 
 | | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, ISOASCORBIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Park, A.K, Kim, H.W. | | Deposit date: | 2016-04-15 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica
Sci Rep, 6, 2016
|
|
5JCL
 
 | | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Os09g0567300 protein | | Authors: | Park, A.K, Kim, H.W. | | Deposit date: | 2016-04-15 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica
Sci Rep, 6, 2016
|
|
5JCK
 
 | | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Os09g0567300 protein | | Authors: | Park, A.K, Kim, H.W. | | Deposit date: | 2016-04-15 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica
Sci Rep, 6
|
|
5JCN
 
 | | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica | | Descriptor: | ASCORBIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Park, A.K, Kim, H.W. | | Deposit date: | 2016-04-15 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica
Sci Rep, 6, 2016
|
|
6IO1
 
 | |
2P7U
 
 | |
2OZ2
 
 | |
1JO7
 
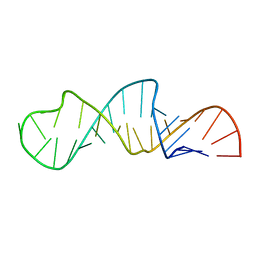 | | Solution Structure of Influenza A Virus Promoter | | Descriptor: | Influenza A virus promoter RNA | | Authors: | Bae, S.-H, Cheong, H.-K, Lee, J.-H, Cheong, C, Kainosho, M, Choi, B.-S. | | Deposit date: | 2001-07-27 | | Release date: | 2001-09-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural features of an influenza virus promoter and their implications for viral RNA synthesis.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7EXP
 
 | | Crystal structure of zebrafish TRAP1 with AMPPNP and MitoQ | | Descriptor: | 2,3-dimethoxy-5-methyl-6-[10-(triphenyl-$l^{5}-phosphanyl)decyl]cyclohexa-2,5-diene-1,4-dione, COBALT (II) ION, MAGNESIUM ION, ... | | Authors: | Lee, H, Yoon, N.G, Kang, B.H, Lee, C. | | Deposit date: | 2021-05-28 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Mitoquinone Inactivates Mitochondrial Chaperone TRAP1 by Blocking the Client Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
7XDP
 
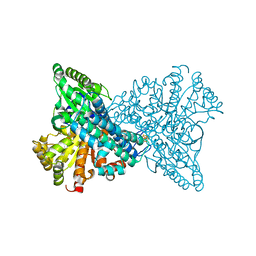 | |
7XDM
 
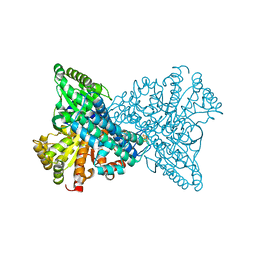 | | ChCODH2 A559W mutant in anaerobic condition | | Descriptor: | Carbon monoxide dehydrogenase 2, FE(4)-NI(1)-S(5) CLUSTER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Heo, Y.Y, Kim, S.M. | | Deposit date: | 2022-03-28 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | O2-tolerant CO dehydrogenase via tunnel redesign for the removal of CO from industrial flue gas
Nat Catal, 5, 2022
|
|
7XDN
 
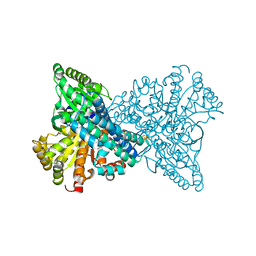 | |
5X9V
 
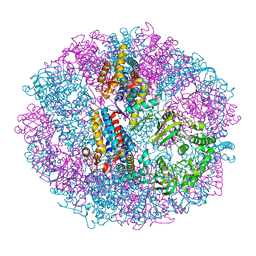 | | Crystal structure of group III chaperonin in the Closed state | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Thermosome, ... | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2017-03-09 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural and mechanistic characterization of an archaeal-like chaperonin from a thermophilic bacterium
Nat Commun, 8, 2017
|
|
