4N3Z
 
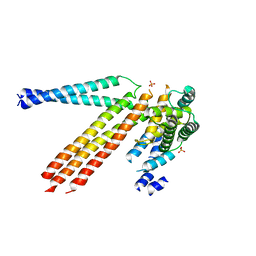 | |
4N3X
 
 | |
5Z10
 
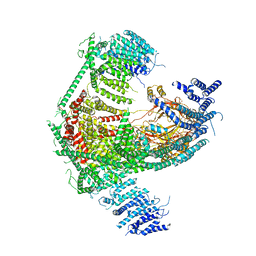 | | Structure of the mechanosensitive Piezo1 channel | | Descriptor: | Piezo-type mechanosensitive ion channel component 1 | | Authors: | Zhao, Q, Zhou, H, Chi, S, Wang, Y, Wang, J, Geng, J, Wu, K, Liu, W, Zhang, T, Dong, M.-Q, Wang, J, Li, X, Xiao, B. | | Deposit date: | 2017-12-22 | | Release date: | 2018-01-31 | | Last modified: | 2020-01-29 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structure and mechanogating mechanism of the Piezo1 channel.
Nature, 554, 2018
|
|
4ROV
 
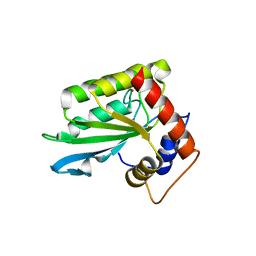 | | The crystal structure of novel APOBEC3G CD2 head-to-tail dimer suggests the binding mode of full-length APOBEC3G to HIV-1 ssDNA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Lu, X, Zhang, T, Xu, Z, Liu, S, Zhao, B, Lan, W, Wang, C, Ding, J, Cao, C. | | Deposit date: | 2014-10-29 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of DNA cytidine deaminase ABOBEC3G catalytic deamination domain suggests a binding mode of full-length enzyme to single-stranded DNA
J.Biol.Chem., 290, 2015
|
|
4ROW
 
 | | The crystal structure of novel APOBEC3G CD2 head-to-tail dimer suggests the binding mode of full-length APOBEC3G to HIV-1 ssDNA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Lu, X, Zhang, T, Xu, Z, Liu, S, Zhao, B, Lan, W, Wang, C, Ding, J, Cao, C. | | Deposit date: | 2014-10-29 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of DNA cytidine deaminase ABOBEC3G catalytic deamination domain suggests a binding mode of full-length enzyme to single-stranded DNA
J.Biol.Chem., 290, 2015
|
|
1WSP
 
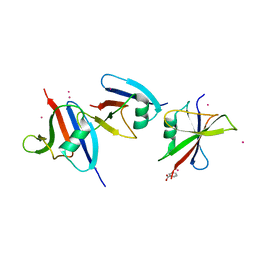 | | Crystal structure of axin dix domain | | Descriptor: | Axin 1 protein, BENZOIC ACID, MERCURY (II) ION | | Authors: | Shibata, N, Hanamura, T, Yamamoto, R, Ueda, Y, Yamamoto, H, Kikuchi, A, Higuchi, Y. | | Deposit date: | 2004-11-08 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of axin dix domain
to be published
|
|
1QEZ
 
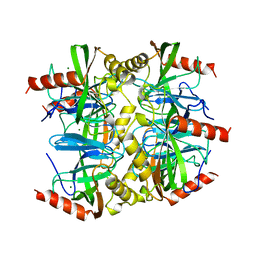 | | SULFOLOBUS ACIDOCALDARIUS INORGANIC PYROPHOSPHATASE: AN ARCHAEL PYROPHOSPHATASE. | | Descriptor: | MAGNESIUM ION, PROTEIN (INORGANIC PYROPHOSPHATASE) | | Authors: | Leppanen, V.-M, Nummelin, H, Hansen, T, Lahti, R, Schafer, G, Goldman, A. | | Deposit date: | 1999-04-06 | | Release date: | 1999-04-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sulfolobus acidocaldarius inorganic pyrophosphatase: structure, thermostability, and effect of metal ion in an archael pyrophosphatase.
Protein Sci., 8, 1999
|
|
3V9R
 
 | | Crystal structure of Saccharomyces cerevisiae MHF complex | | Descriptor: | SULFATE ION, Uncharacterized protein YDL160C-A, Uncharacterized protein YOL086W-A | | Authors: | Yang, H, Zhang, T, Zhong, C, Li, H, Zhou, J, Ding, J. | | Deposit date: | 2011-12-28 | | Release date: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Saccharomyces Cerevisiae MHF Complex Structurally Resembles the Histones (H3-H4)(2) Heterotetramer and Functions as a Heterotetramer
Structure, 20, 2012
|
|
7VKR
 
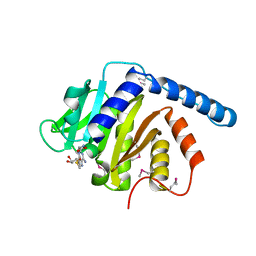 | |
7VKQ
 
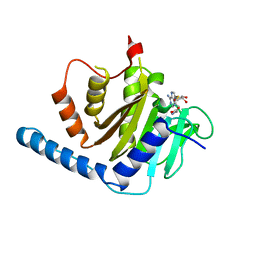 | |
7F00
 
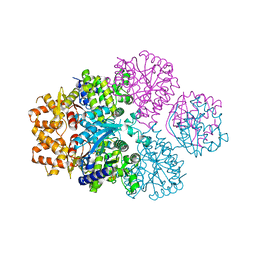 | | Crystal structure of SPD_0310 | | Descriptor: | SULFATE ION, UPF0371 protein SPRM200_0309 | | Authors: | Cao, K, Zhang, T, Li, N, Yang, X, Ding, J, He, Q, Sun, X. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification and Tetramer Structure of Hemin-Binding Protein SPD_0310 Linked to Iron Homeostasis and Virulence of Streptococcus pneumoniae.
Msystems, 7, 2022
|
|
7WQL
 
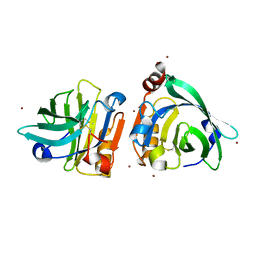 | | Bovin Beta-lactoglobulin binding with zinc ions | | Descriptor: | Beta-lactoglobulin, ZINC ION | | Authors: | Li, T, Ma, J, Zang, J, Zhao, G, Zhang, T. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Zinc binding strength of proteins dominants zinc uptake in Caco-2 cells.
Rsc Adv, 12, 2022
|
|
7WQG
 
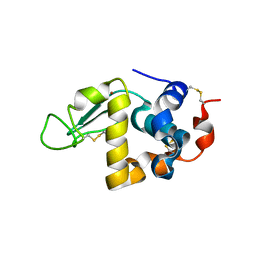 | | Bovin Alpha-lactalbumin binding with zinc ions | | Descriptor: | Alpha-lactalbumin, ZINC ION | | Authors: | Li, T, Zhang, T. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Zinc binding strength of proteins dominants zinc uptake in Caco-2 cells.
Rsc Adv, 12, 2022
|
|
4HR6
 
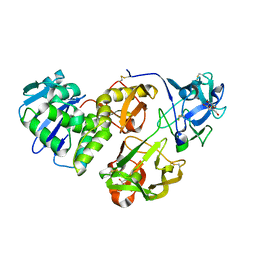 | | Crystal structure of snake gourd (Trichosanthes anguina) seed lectin, a three chain homologue of type II RIPs | | Descriptor: | LECTIN, methyl alpha-D-galactopyranoside | | Authors: | Sharma, A, Pohlentz, G, Bobbili, K.B, Jeyaprakash, A.A, Chandran, T, Mormann, M, Swamy, M.J, Vijayan, M. | | Deposit date: | 2012-10-26 | | Release date: | 2013-08-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The sequence and structure of snake gourd (Trichosanthes anguina) seed lectin, a three-chain nontoxic homologue of type II RIPs.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1LA3
 
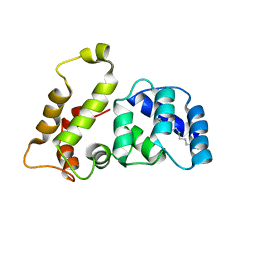 | | Solution structure of recoverin mutant, E85Q | | Descriptor: | CALCIUM ION, MYRISTIC ACID, Recoverin | | Authors: | Ames, J.B, Hamasaki, N, Molchanova, T. | | Deposit date: | 2002-03-27 | | Release date: | 2002-06-19 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structure and calcium-binding studies of a recoverin mutant (E85Q) in an allosteric intermediate state.
Biochemistry, 41, 2002
|
|
4LTU
 
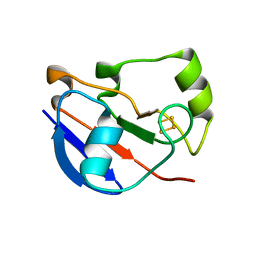 | | Crystal Structure of Ferredoxin from Rhodopseudomonas palustris HaA2 | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin | | Authors: | Zhou, W.H, Zhang, T, Yang, H, Bell, S.G, Wong, L.-L. | | Deposit date: | 2013-07-24 | | Release date: | 2014-10-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of Ferredoxin from Rhodopseudomonas palustris HaA2
To be Published
|
|
3L5A
 
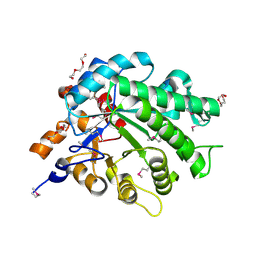 | | Crystal structure of a probable NADH-dependent flavin oxidoreductase from Staphylococcus aureus | | Descriptor: | NADH/flavin oxidoreductase/NADH oxidase, TRIETHYLENE GLYCOL | | Authors: | Lam, R, Gordon, R.D, Vodsedalek, J, Battaile, K.P, Grebemeskel, S, Lam, K, Romanov, V, Chan, T, Mihajlovic, V, Thompson, C.M, Guthrie, J, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2009-12-21 | | Release date: | 2010-12-22 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a probable NADH-dependent flavin oxidoreductase from Staphylococcus aureus
To be Published
|
|
3UIU
 
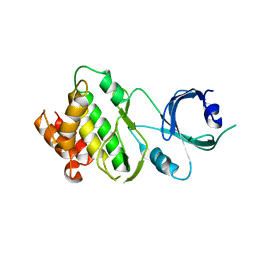 | | Crystal structure of Apo-PKR kinase domain | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase | | Authors: | Li, F, Li, S, Yang, X, Shen, Y, Zhang, T. | | Deposit date: | 2011-11-06 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Crystal structure of Apo-PKR kinase domain
TO BE PUBLISHED
|
|
4M6W
 
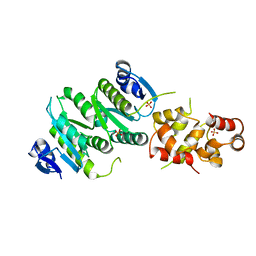 | | Crystal structure of the C-terminal segment of FANCM in complex with FAAP24 | | Descriptor: | Fanconi anemia group M protein, Fanconi anemia-associated protein of 24 kDa, SULFATE ION | | Authors: | Yang, H, Zhang, T, Tong, L, Ding, J. | | Deposit date: | 2013-08-11 | | Release date: | 2013-10-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the functions of the FANCM-FAAP24 complex in DNA repair.
Nucleic Acids Res., 41, 2013
|
|
7X9E
 
 | | Crystal structure of the 76E1 Fab in complex with a SARS-CoV-2 spike peptide | | Descriptor: | 76E1 Fab Heavy Chain, 76E1 Fab Light Chain, Spike peptide | | Authors: | Chen, X, Zhang, T, Ding, J, Sun, X, Sun, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Neutralization mechanism of a human antibody with pan-coronavirus reactivity including SARS-CoV-2.
Nat Microbiol, 7, 2022
|
|
3L20
 
 | | Crystal structure of a hypothetical protein from Staphylococcus aureus | | Descriptor: | Putative uncharacterized protein | | Authors: | Lam, R, Chan, T, Battaile, K.P, Mihajlovic, V, Romanov, V, Soloveychik, M, Kisselman, G, McGrath, T.E, Lam, K, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2009-12-14 | | Release date: | 2010-10-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Crystal structure of a hypothetical protein from Staphylococcus aureus
To be Published
|
|
4OZR
 
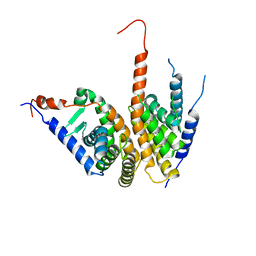 | | Crystal structure of the ligand binding domains of the Bovicola ovis ecdysone receptor EcR/USP heterodimer (methylene lactam crystal) | | Descriptor: | Ecdysone receptor, Retinoid X receptor | | Authors: | Ren, B, Peat, T.S, Streltsov, V.A, Pollard, M, Fernley, R, Grusovin, J, Seabrook, S, Pilling, P, Phan, T, Lu, L, Lovrecz, G.O, Graham, L.D, Hill, R.J. | | Deposit date: | 2014-02-18 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unprecedented conformational flexibility revealed in the ligand-binding domains of the Bovicola ovis ecdysone receptor (EcR) and ultraspiracle (USP) subunits.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1ZTX
 
 | | West Nile Virus Envelope Protein DIII in complex with neutralizing E16 antibody Fab | | Descriptor: | Envelope protein, Heavy Chain of E16 Antibody, Light Chain of E16 Antibody | | Authors: | Nybakken, G.E, Oliphant, T, Diamond, M.S, Fremont, D.H. | | Deposit date: | 2005-05-27 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of West Nile virus neutralization by a therapeutic antibody.
Nature, 437, 2005
|
|
2N9L
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for in-cell GB1 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ikeya, T, Hanashima, T, Hosoya, S, Shimazaki, M, Ikeda, S, Mishima, M, Guentert, P, Ito, Y. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved in-cell structure determination of proteins at near-physiological concentration
Sci Rep, 6, 2016
|
|
2N9K
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for in vitro GB1 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ikeya, T, Hanashima, T, Hosoya, S, Shimazaki, M, Ikeda, S, Mishima, M, Guentert, P, Ito, Y. | | Deposit date: | 2015-11-26 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved in-cell structure determination of proteins at near-physiological concentration
Sci Rep, 6, 2016
|
|
